Subfamily: Alpharhabdovirinae
Genus: Almendravirus
Distinguishing features
Viruses assigned to the genus Almendravirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Almendraviruses are capable of replication in mosquito cells in vitro but appear to be unable to replicate in vertebrates and so may infect only mosquitoes. The genome organisation features a gene encoding a small class 1a viroporin-like protein between the G and L genes.
Virion
Morphology
In ultra-thin sections of infected insect cells, bullet-shaped virions (130–460 nm × 40–55 nm) have been reported but these may include defective-interfering particles and longitudinally fused virions (Vasilakis et al., 2014, Contreras et al., 2017).
Nucleic acid
Almendravirus genomes consist of a single molecule of negative-sense, single-stranded RNA and range from approximately 10.8–11.9 kb (Vasilakis et al., 2014, Contreras et al., 2017).
Proteins
The N, P, M, G and L share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses. The small class 1a viroporin-like proteins encoded in the U1 gene range from 51 to 80 amino acids (6.1–9.5 kDa) and feature a very short N-terminal domain, a predicted hydrophobic transmembrane domain and a C-terminal domain that is rich in basic residues (Vasilakis et al., 2014, Contreras et al., 2017).
Genome organisation and replication
Almendravirus genomes include five genes (N, P, M, G and L) encoding the structural proteins and an additional gene between the G and L genes encoding a class 1a viroporin-like protein (U1) (Vasilakis et al., 2014, Contreras et al., 2017). In the Balsa virus (species Almendravirus balsa) genome, there is an alternative ORF (Px) within the P gene. All other almendravirus ORFs occur in discrete transcriptional units including conserved transcription initiation and transcription termination/polyadenylation sequences (Figure 1 Almendravirus).
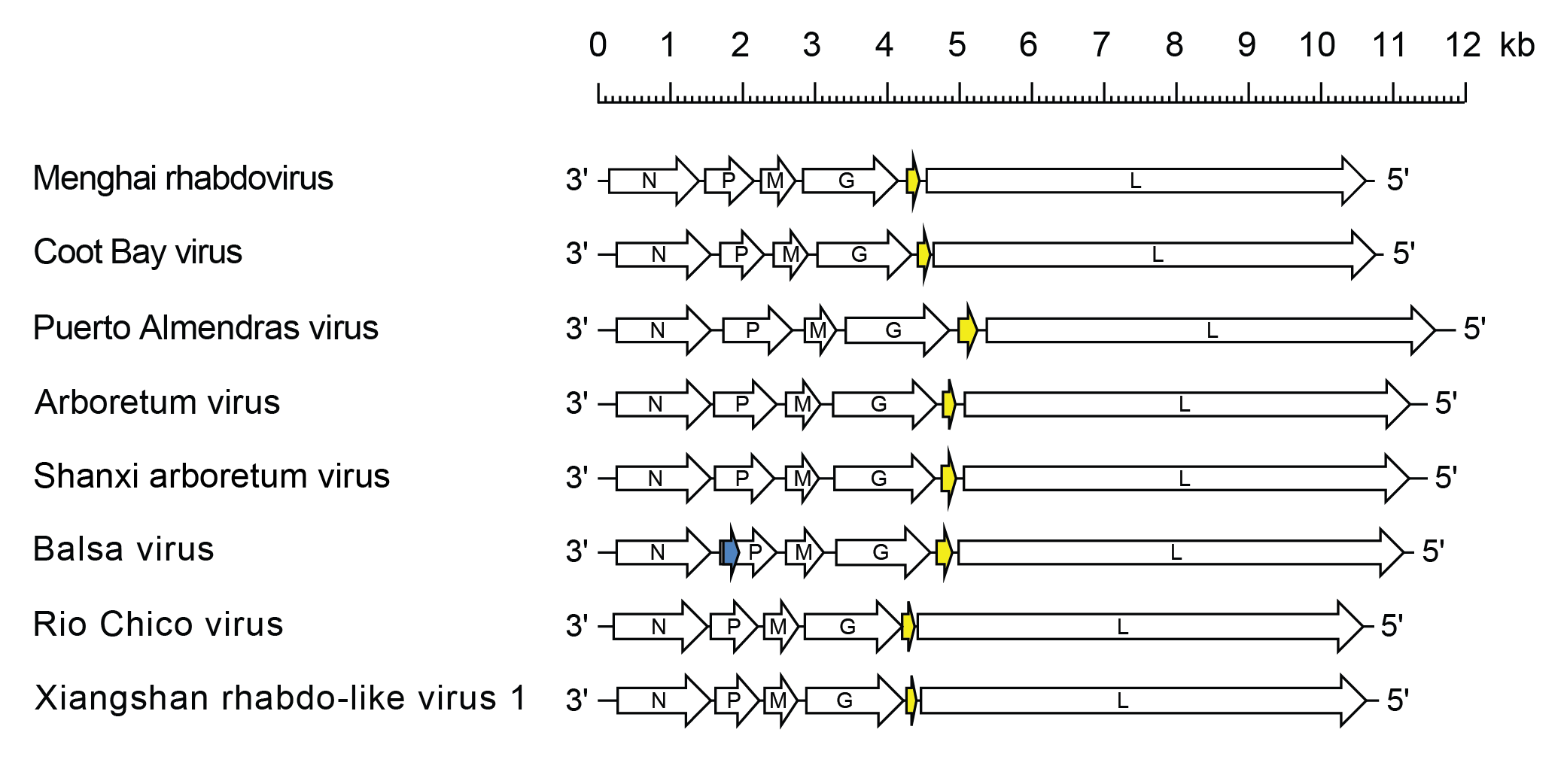 |
| Figure 1 Almendravirus. Schematic representation of almendravirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. The U1 ORFs (yellow) each encode class 1a viroporin-like proteins. An alternative ORF (Px) in the P gene of Balsa virus is also shown (blue). |
Biology
Each of the viruses assigned to the genus was either isolated from mosquitoes or detected in insect pools that included unidentified dipteran insects. Several viruses replicate in mosquito cell lines but failed to replicate in vertebrate (mammal, bird or reptile) cell cultures or in suckling mice. They have been isolated from Culex, Aedes, Anopheles and Psorophora mosquitoes and may be mosquito-specific viruses. The known distribution of members of the genus includes Peru, Panama, Colombia, Florida and China (Vasilakis et al., 2014, Contreras et al., 2017).
Species demarcation criteria
Viruses assigned to different species within the genus Almendravirus have several of the following characteristics: A) minimum amino acid sequence divergence of 5% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) can be distinguished in virus neutralisation tests; and F) occupy different ecological niches as evidenced by differences in hosts and/or arthropod vectors.

