Subfamily: Alpharhabdovirinae
Genus: Siniperhavirus
Distinguishing features
Viruses assigned to the genus Siniperhavirus belong to one of the five genera of rhabdoviruses that infect finfish, the other genera being Perhabdovirus, Scophrhavirus, Sprivivirus and Novirhabdovirus. Viruses assigned to the genus Siniperhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. They are most closely related to perhabdoviruses, scophrhaviruses and cetarhaviruses (infecting cetaceans).
Virion
Morphology
Siniperhavirus virions have bullet-shaped morphology. Various dimensions have been reported in the range 110–500 nm in length and 60–200 nm in diameter (Tao et al., 2007, Axén et al., 2017, Liu et al., 2019, Lyu et al., 2019).
Nucleic acid
The genome consists of a single molecule of negative-sense, single-stranded RNA of approximately 11.0–11.5 kb (Tao et al., 2007, Zeng et al., 2014, Liu et al., 2015, Axén et al., 2017, Liu et al., 2019, Lyu et al., 2019).
Proteins
N, P, M, G and L share significant sequence homology with the homologous proteins of other rhabdoviruses.
Genome organisation and replication
Siniperhavirus genomes include five genes in the order 3′-N-P-M-G-L-5′ encoding a nucleoprotein, polymerase-associated protein, matrix protein, glycoprotein and RNA-directed RNA polymerase, respectively (Figure 1 Siniperhavirus). In Siniperca chuatsi rhabdovirus (SCRV; species Siniperhavirus chuatsi), there is an alternative ORF (Px) in the P gene commencing upstream of the P ORF and encoding a putative basic protein of 8.5 kDa (Zhu et al., 2011). An additional small ORF following the M ORF in the SCRV M gene is not conserved in all isolates of the virus and so is unlikely to be expressed functionally (Tao et al., 2008). The SCRV genome contains a leader region of 71 nt preceding the transcription initiation of the N gene, and a trailer of 61 nt following the transcription termination of the L gene (Zhu et al., 2011). The transcriptional initiation and the termination/polyadenylation signals are conserved for all genes, 3′-UUGUG and 3′-AUAC(U)7, respectively, but the non-transcribed intergenic regions are variable. There is inverse complementarity between the 3′-leader and 5′-trailer sequences.
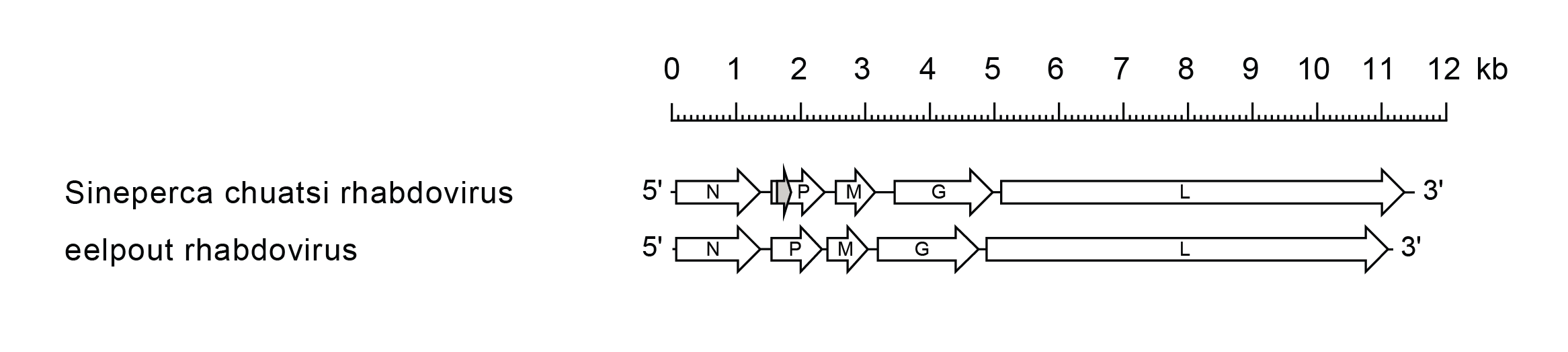 |
| Figure 1 Siniperhavirus. Schematic representation of siniperhavirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. There is an alternative ORF (Px) in the SCRV P gene (grey). |
Biology
Eelpout rhabdovirus (EPRV; species Siniperhavirus zoarces) was detected by high-throughput sequencing in samples of eelpout (Zoarces viviparous) collected during mass fish mortalities near Stockholm, Sweden (Zoarces viviparous) in the Baltic Sea, J Fish Dis, 40, 2, 219-29">Axén et al., 2017). SCRV was first isolated from mandarin fish (Siniperca chuatsi) collected in Guangdong Province, China, in 1999 (Zhang and Li 1999, Zhang et al., 2005, Tao et al., 2007) and subsequently from diseased largemouth bass (Micropterus salmoides) from the same province in 2011 (Ma et al., 2013, Zeng et al., 2014). Several other viruses reported from China share very high levels of nucleotide and amino acid sequence identity with SCRV and can be considered isolates of the same virus. These include hybrid snakehead rhabdovirus which was isolated on several occasions from moribund snakehead fish or hybrids (Channa maculata x Channa argus) collected in Guangdong Province and was shown experimentally to also infect mandarin fish (Liu et al., 2015); Chinese rice-field eel rhabdovirus which was isolated from diseased Asian swamp eels (Monopterus albus) collected from Hubei Province (Liu et al., 2019); and Micropterus salmoides rhabdovirus which was isolated from moribund largemouth bass (M. salmoides) collected in Zhejiang Province (Lyu et al., 2019). These data indicate that SCRV is a significant pathogen affecting farmed finfish of a wide range of species.
Antigenicity
Polyclonal mouse antiserum to SCRV failed to cross-react in immunoblots with the N proteins of Scophthalmus maximus rhabdovirus (genus Scohprhavirus) or spring viraemia of carp virus (genus Sprivvirus) (Tao et al., 2007).
Species demarcation criteria
Viruses assigned to different species within the genus Siniperhavirus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in N proteins; B) minimum sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in G proteins; D) can be distinguished in virus neutralisation tests; and E) occupy different ecological niches as evidenced by differences in vertebrate hosts.

