Subfamily: Alpharhabdovirinae
Genus: Lyssavirus
Distinguishing features
Viruses assigned to the genus Lyssavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Lyssaviruses cause acute progressive encephalomyelitis (rabies) in mammals, being transmitted between susceptible individuals directly by bites, scratches or contamination of mucous membranes with infected saliva, without participation of arthropod vectors. Bats (order Chiroptera) are the principal reservoir hosts for most lyssaviruses, whereas carnivores (order Carnivora), as well as bats, maintain circulation of rabies virus (RABV; species Lyssavirus rabies). Viruses assigned to the genus are distributed worldwide, except Antarctica and several isolated islands, although viruses of different species have different circulation ranges, and are typically delineated by broad geographical clustering of phylogenetic groups (Troupin et al., 2016). Lyssavirus genomes contain only five structural protein genes (3′-N-P-M-G-L-5′) but feature a long 3′-untranslated region following the G mRNA.
Virion
Morphology
The virions are bullet-shaped (60–110 nm × 130–250 nm), and composed of two structural units: an internal helical nucleocapsid, approximately 50 nm in diameter, and a lipid envelope which is derived from the host cytoplasmic membrane during budding (Hummeler et al., 1967). Knobbed spikes (8 nm in length), consisting of three glycosylated ectodomains of G, protrude through the virion membrane (Gaudin et al., 1992).
Physicochemical and physical properties
RABV particles consist of RNA (2–3%), protein (67–74%), lipid (20–26%) and carbohydrate (3%).
Nucleic acid
The single molecule of negative-sense single-stranded RNA is approximately 11.9–12.3 kb in length. The RNA is tightly associated with N within the RNP.
Proteins
The five major polypeptides include: N (Mr = 58–62 kDa), P (Mr = 35–40 kDa), M (Mr = 22–25 kDa), G (Mr = 65–80 kDa), and L (Mr = 190 kDa). RABV G is glycosylated and palmitoylated at sites that have been mapped (Wunner et al., 1985, Gaudin et al., 1991). N and P are both phosphoproteins and, in the case of RABV, phosphorylation involves different host protein kinases: cellular casein kinase II is implicated for N; and P is phosphorylated by certain isomers of protein kinase C as well as by an additional kinase, RVPK, yet to be clearly defined (Gupta et al., 2000). The N:P ratio in the RNP of RABV is 2:1 per virion, which indicates that two molecules of N interact with 1 molecule of P. M is not phosphorylated; it interacts with both RNP and G in virion assembly to form bullet-shaped virions (Mebatsion et al., 1999). In infected cells, RABV N, P and M also have roles in modulating the host response to infection. N has been shown to inhibit retinoic acid-inducible gene I (RIG-I)-mediated activation of type 1 interferon (IFN) (Masatani et al., 2010, Masatani et al., 2013). Several isoforms of the RABV P (P1–P5) are expressed from five in-frame initiation codons (Chenik et al., 1995) and function to inhibit both interferon (IFN) induction and IFN-dependent signalling (Brzozka et al., 2005, Brzozka et al., 2006) by suppressing interferon regulatory factor 3 (IRF-3) and binding to signal transducers and activator of transcription 1 (STAT1) and STAT2 and interacting with Janus kinase 1 (JAK1) (Vidy et al., 2005, Brzozka et al., 2006, Vidy et al., 2007, Rieder et al., 2011, Sonthonnax et al., 2019). M, in cooperation with P, modulates apoptosis (Kassis et al., 2004) and interacts with the necrosis factor (NF)-κB pathway to inhibit induction of IFN-β, JAK1 and STAT1 (Luco et al., 2012, Ben Khalifa et al., 2016, Sonthonnax et al., 2019). The binding of viral proteins to the PDZ-binding domains of host proteins could also represent a virulence marker (Caillet-Saguy et al., 2015). The envelope contains other host-derived minor protein components. A lipoprotein bilayer consists of a mixture of host-derived lipids, including phospholipids, neutral lipids and glycolipids.
Genome organisation and replication
The RABV genome consists of a 58 nt leader sequence, followed by five structural protein genes in the order 3′-N-P-M-G-L-5′, separated by non-transcribed intergenic regions and followed by a 57–70 nt trailer (Kuzmin et al., 2008) (Figure 1 Lyssavirus). Transcription initiation signal of each mRNA is conserved (3′-UUGUXR-5′), as well as the transcription termination-polyadenylation signal (3′-WCUUUUUUU-5′). Untranslated regions of mRNAs are relatively short, except the 3′-UTR of the G mRNA, which is approximately 440–700 nt in length. In West Caucasian bat virus (WCBV; species Lyssavirus caucasicus), the long 3′-region of the G mRNA contains an ORF of 180 nt (60 amino acids) but it is not known if the corresponding proteins are expressed in infected cells (Kuzmin et al., 2008, Walker et al., 2015). Intergenic regions are variable (2–100 nt), but their lengths increase in the 3′-to-5′ direction, which has the potential effect of causing a progressive slowing and decreasing efficiency of transcription. RABV transcription and replication occur in the cytoplasm of infected cells and follow processes that are similar to those described for vesiculoviruses and other rhabdoviruses (Albertini et al., 2011).
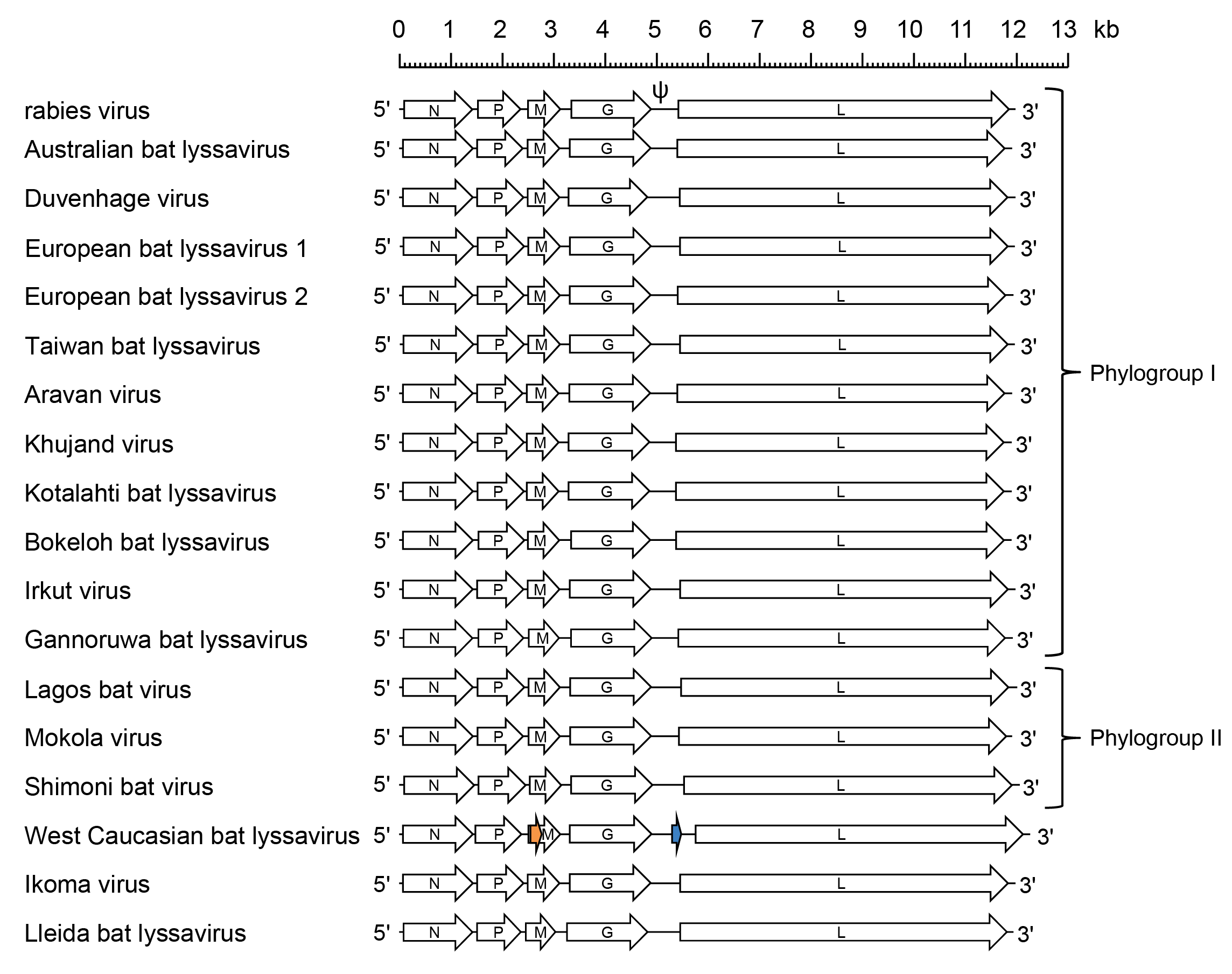 |
| Figure 1 Lyssavirus. Schematic representation of lyssavirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. The genomes feature long untranslated regions in the G genes (designated ψ in rabies virus); in West Caucasian bat virus this ~700 nt region contains a 180 nt ORF (blue) encoding a possible 7.1 kDa protein. West Caucasian bat virus also features an alternative ORF of 228 nt (orange), that commences near the start of the M gene, encoding a possible 8.4 kDa protein. It is not known if these proteins are expressed in infected cells. |
Biology
Lyssaviruses are essential neurotropic pathogens (Fooks et al., 2017). Delivered into a wound via a bite or wound contamination, the virus can replicate at the inoculation site, as was shown for skeletal muscle cells (Murphy and Bauer 1974). The virus then reaches the motor or sensory neurons and propagates up to the central nervous system (CNS) by following neuronal connections and using retrograde axonal transport (Jackson 2013). Neuronal pathways shield the virus from host immune surveillance, resulting in absence of early antibody response (Johnson et al., 2010). Being delivered to the CNS, the virus disseminates rapidly. Nearly all regions of the CNS may be affected and RABV has been used to map neuronal connections (Ugolini 2011). The duration of the asymptomatic incubation period can be variable (two months on average), while the symptomatic clinical period is rapid and severe (about one week) (Udow et al., 2013). Neuropathological changes observed in the infected brain are relatively mild histologically and include gliosis, slight neuronophagia, perivascular infiltration with inflammatory cells, with rare involvement of meninges (Jackson 2013). Occasionally, more severe brain damage occurs, such as spongiform lesions, extensive neuronal degeneration and widespread inflammation (Jackson 2013). Functional alteration of the CNS is much more significant than is morphological representation. Reverse dissemination of virus from the CNS during the clinical period of rabies occurs along peripheral nerves. Viral RNA may be detected in a variety of organs and tissues at the end of the clinical period. However, only low titres of infectious virus occasionally can be isolated from extraneural tissues (Jackson et al., 1999). The exception is the salivary glands, in which virus undergoes additional replication cycles and is released into the saliva to complete the transmission (Jackson 2013). Acute generalised CNS dysfunction leads to a lethal outcome of the disease. Very rare cases of survival after manifestation of clinical signs of rabies have been observed in humans and some animals (Jackson et al., 2003). However, these sporadic events cannot be taken as support for a theory of lyssavirus persistence.
In nature, lyssaviruses are associated with specific mammalian reservoir hosts, predominantly bats and carnivores. Spill-over infections into vertebrates of other species lead to a dead end of the transmission chain in the vast majority of cases. The exception is RABV, which is distributed most broadly, and host shifts of certain variants, with the establishment of sustained circulation in a new host of a different species, have been well documented (Badrane and Tordo 2001, Rupprecht et al., 2011, Kuzmin et al., 2012, Troupin et al., 2016, Marston et al., 2017b, Marston et al., 2018). Laboratory rodents (mice, hamsters), foxes and bats are highly susceptible to intra-cranial inoculation with lyssaviruses, whereas their susceptibility to peripheral inoculation varies, depending on viral species and lineage, inoculation dose and route (Fekadu et al., 1988a, Vos et al., 2004, Eggerbauer et al., 2017). Cell cultures derived from the mammalian nervous tissue (such as mouse neuroblastoma, MNA cells) are more susceptible to lyssaviruses than are other mammalian cell cultures (BHK, Vero, etc.). Successful propagation in insect cell lines has been described only for Mokola virus (MOKV; species Lyssavirus mokola) (Buckley 1975).
Antigenicity
Based on antigenic properties and phylogenetic relationships, viruses in the genus have been subdivided into two phylogroups (Figure 1.Lyssavirus). Phylogroup I includes RABV, Australian bat lyssavirus (ABLV; species Lyssavirus australis), Duvenhage virus (DUVV; species Lyssavirus duvenhage), European bat lyssavirus 1 (EBLV-1; species Lyssavirus hamburg), European bat lyssavirus 2 (EBLV-2; species Lyssavirus helsinki), Aravan virus (ARAV; species Lyssavirus aravan), Khujand virus (KHUV; species Lyssavirus khujand), Kotalathi bat lyssavirus (KBLV; species Lyssavirus kotalahti), Bokeloh bat lyssavirus (BBLV; species Lyssavirus bokeloh), Irkut virus (IRKV; species Lyssavirus irkut), Taiwan bat lyssavirus (TWBLV; species Lyssavirus formosa) and Gannoruwa bat lyssavirus (GBLV; species Lyssavirus gannoruwa); phylogroup II includes MOKV, Lagos bat virus (LBV; species Lyssavirus lagos) and Shimoni bat virus (SHIBV; species Lyssavirus shimoni) (Badrane et al., 2001, Fooks 2004, Gunawardena et al., 2016, Calvelage et al., 2021). The most divergent viruses in the genus, WCBV, Ikoma lyssavirus (IKOV; species Lyssavirus ikoma) and Lleida bat lyssavirus (LLBV; species Lyssavirus lleida), are not members of either of these phylogroups (Weyer et al., 2008, Marston et al., 2012, Horton et al., 2014, Marston et al., 2017a).
Antigenic cross-reactivity correlates with the relatively short genetic distances between lyssaviruses. Antigens of the RNP, which are most abundant in infected cells, cross-react between all members of the genus described to date (Rupprecht et al., 1991). This feature facilitates the use of standardised diagnostic reagents for detection of all lyssaviruses (e.g. by direct fluorescent antibody or immunohistochemical assays). In contrast, glycoprotein antigens are relatively conserved within phylogroups (ectodomain conservation >75%) but not between phylogroups (ectodomain conservation <65%). As a result, commercially available rabies vaccines and anti-rabies immunoglobulins, that mainly induce or provide neutralising antibodies targeting the glycoprotein, protect against phylogroup I lyssaviruses but not against other lyssaviruses (Fekadu et al., 1988b, Bahloul et al., 1998, Badrane et al., 2001, Fooks 2004, Hanlon et al., 2005).
Species demarcation criteria
Genetic distances between members of different lyssavirus species are significantly shorter than the distances between viruses of different species in other rhabdovirus genera, which has been attributed to evolutionary constraints, possibly imposed by their unique pathobiology or their vector/reservoir preferences.
Viruses assigned to different species within the genus Lyssavirus have several of the following characteristics: A) Genetic distances, with the threshold of 78-80% nt identity for the complete N gene, provides better quantitative resolution compared to other genes, or 80% nt identity for concatenated coding regions of N+P+M+G+L; B) In phylogenetic trees based on the entire N, or G, or L gene sequences, or concatenated N+P+M+G+L coding sequences, the new virus does not represent a sister branch to a virus from an established species. Instead, it is placed ancestrally to a group (cluster) of phylogenetically-related viruses that belong to several established species; C) Can be distinguished serologically in virus-neutralization tests; D) Occupies a distinct ecologic niche as evidenced by host species, pathobiological properties, or geographical range.
Related, unclassified viruses
| Virus name | Accession number
| Virus abbreviation
|
| Divaea bat lyssavirus | OQ428158 | DBLV |
| Phala bat lyssavirus | OQ970171 | PBLV |
| Taiwan bat lyssavirus 2 | ON437589 | TWBLV2 |
Virus names and virus abbreviations are not official ICTV designations.

