Subfamily: Deltarhabdovirinae
Genus: Gammaricinrhavirus
Distinguishing features
Viruses assigned to the genus Gammaricinrhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Members of the genus have been detected in hard ticks (family Ixodidae). They are distinct phylogenetically from viruses assigned to the genera Alpharicinrhavirus and Betaricinrhavirus.
Virion
Morphology
Virion morphology is unknown.
Nucleic acid
The genomes consist of a single molecule of negative-sense, single-stranded RNA of approximately 13.4–13.7 kb (Li et al., 2015, Kong et al., 2022).
Proteins
The N, P, M, G and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses.
Genome organisation and replication
The genomes include the five canonical rhabdovirus structural protein genes (N, P, M, G and L) and one additional gene (U1) between the M and G genes encoding a protein of approximately 35 kDa (Figure 1 Gammaricinrhavirus).
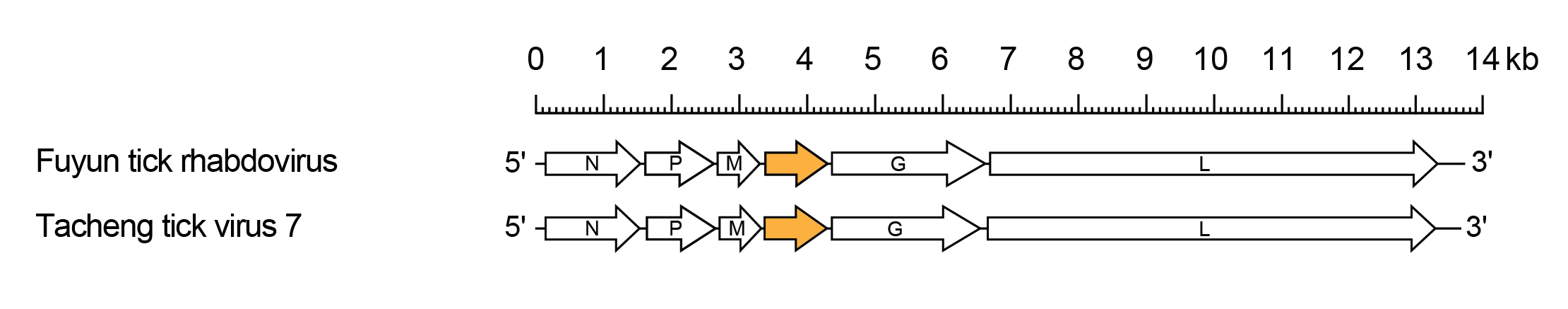 |
| Figure 1 Gammaricinrhavirus. Schematic representation of the gammahymrhavirus genomes shown in reverse (positive-sense) polarity. The genomes contain long open reading frames (ORFs) in the N, P, M, G and L genes (open arrows) and an additional ORFs between M and G (orange) |
Biology
The viruses assigned to this genus were discovered by viral metagenomic sequencing of hard ticks (Ixodidae) from China. Fuyun tick rhabdovirus (species Gammaricinrhavirus fuyun) was detected in Hyalomma sp. ticks collected in Fuyun, Xinjiang Province (Kong et al., 2022). Tacheng tick virus 7 (species Gammaricinrhavirus tacheng) was detected in Argas miniatus ticks collected in Tacheng, Xinjiang Province (Li et al., 2015). No isolates of the viruses have yet been reported.
Species demarcation criteria
Viruses assigned to different species within the genus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) they can be distinguished in virus neutralisation tests; and F) they occupy different ecological niches as evidenced by differences in vertebrate hosts and or arthropod vectors.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Longquan bat rhabdovirus 1 | OQ715712* | LqBRV1 |
| Wufeng shrew rhabdovirus 11 | OQ715732* | WfSRV11 |
| Wufeng shrew rhabdovirus 16 | OQ715706* | WfSRV16 |
| Wufeng shrew rhabdovirus 17 | OQ715721* | WfSRV17 |
Virus names and virus abbreviations are not official ICTV designations.
* Coding region sequence incomplete

