Subfamily: Alpharhabdovirinae
Genus: Tupavirus
Distinguishing features
Viruses assigned to the genus Tupavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Tupaviruses infect mammals and birds. The genome organisation features an alternative long ORF (Px or C) in the P gene and a gene encoding a small hydrophobic protein (U1 or SH) between the M gene and the G gene.
Virion
Morphology
Both bullet-shaped virions and bacilliform virions (i.e., two rounded ends) have been reported for tupaviruses. For Klamath virus (KLAV; species Tupavirus klamath) and tupaia rhabdovirus (TUPV; species Tupavirus tupaia), enveloped bullet-shaped virions (125–220 nm × 68–80 nm) with helical cores have been observed in cell culture supernates or purified virus preparations under negative stain (Murphy et al., 1972a, Kurz et al., 1986). Surface projections (8–11 nm) forming a regular reticular pattern were also reported. In ultra-thin sections of infected cells (KLAV, TUPV and Durham virus [DURV; species Tupavirus durham]), both bullet-shaped and bacilliform virions have been observed budding from intracytoplasmic membranes and at the cell surface (Murphy et al., 1972a, Kurz et al., 1986, Allison et al., 2011).
Nucleic acid
Tupavirus genomes consist of a single molecule of negative-sense, single-stranded RNA and range from approximately 11.1–12.2 kb (Springfeld et al., 2005, Allison et al., 2011, Walker et al., 2015).
Proteins
The N, P, M, G and L share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses. Other proteins are encoded in the tupavirus genome but have not yet been identified in infected cells. The Px (or C) ORFs, present in all tupaviruses, encode highly basic proteins of 136 to 221 amino acids (15.7–25.8 kDa) (Springfeld et al., 2005, Allison et al., 2011, Walker et al., 2015). The U1 (or SH) gene, also present in all tupaviruses, encodes predicted small hydrophobic proteins of 77 to 93 amino acids (9.2–10.8 kDa). The U2 genes of KLAV and Wufeng Rhinolophus pearsonii tupavirus 1 (WRpeTV1; species Tupavirus pearsonii), tupavirus SB8301 (TUPVSB; species Tupavirus incomtus), bat tupavirus BS1 (BtTVBS1; species Tupavirus stheno) and bat tupavirus BS2 (BtTVBS2; species Tupavirus stoliczkanus) encode very small proteins (6.4–6.5 kDa) (Walker et al., 2015).
Genome organisation and replication
Tupavirus genomes include five genes (N, P, M, G and L) encoding the structural proteins and multiple additional long ORFs (Figure 1 Tupavirus) (Springfeld et al., 2005, Allison et al., 2011, Walker et al., 2015). The genomes of all tupaviruses feature an alternative long ORF (Px or C) in the P gene and an additional gene (U1 or SH) between the M gene and G gene. The genomes of KLAV, WRpeTV1, TUPVSB, BtTVBS1 and BtTVBS2 also features an additional gene (U2) located between the G gene and L gene. The U1 and U2 genes are organised as independent transcriptional units including conserved transcription initiation and transcription termination/polyadenylation sequences, indicating that they are highly likely to be expressed in infected cells.
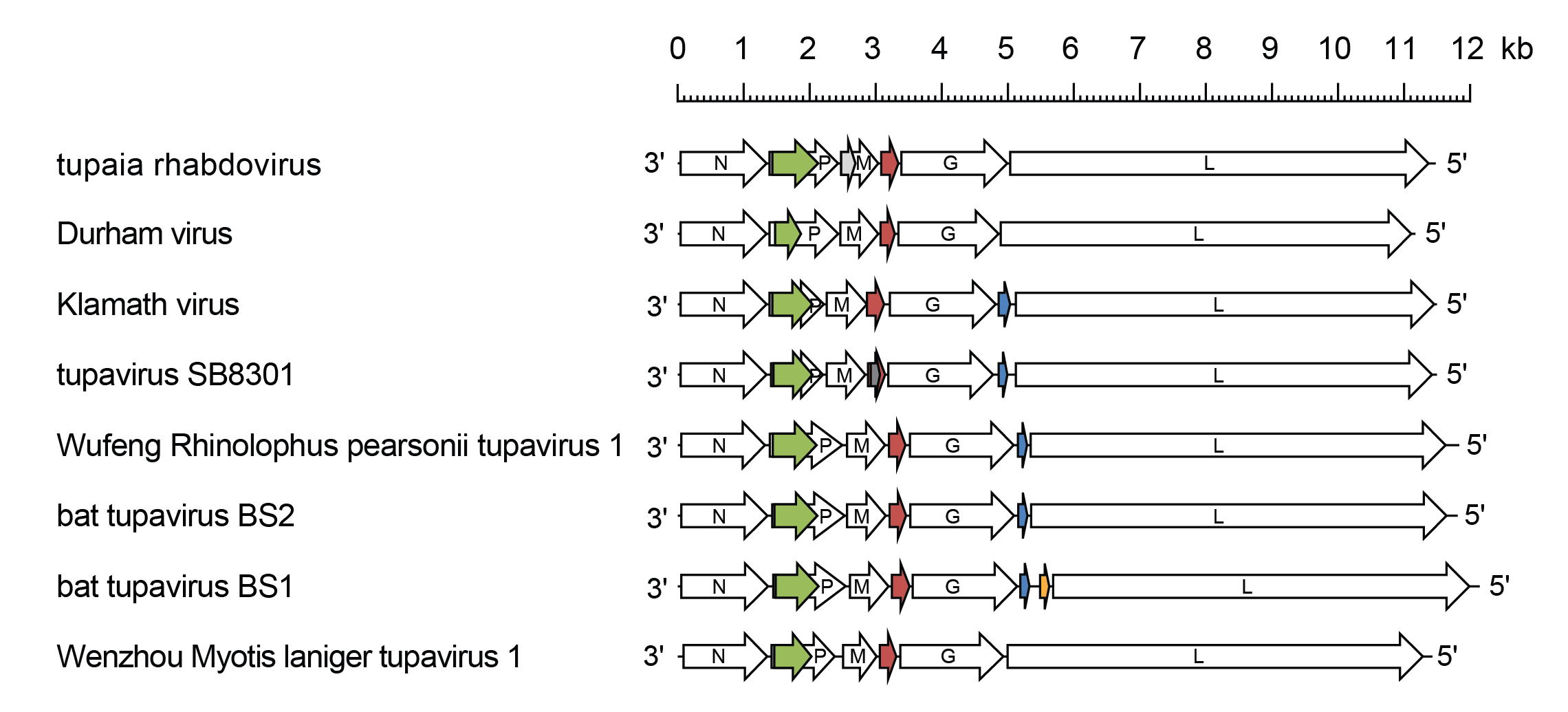 |
| Figure 1 Tupavirus. Schematic representation of tupavirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. ORF Px (green) is in an alternative reading frame in the P gene; ORF U1 (red brown), ORF U2 (blue) and ORF U3 (orange) are in independent transcriptional units bounded by consensus transcription initiation and transcription termination/polyadenylation sequences. Other small ORFs (≥150 nt or 50 aa) occur in alternative reading frames (grey) in tupaia rhabdovirus and tupavirus SB8301 virus but may or may not be expressed. |
Biology
Tupaviruses infect various birds and mammals. TUPV was isolated in Germany from liver tumour cells removed from a northern tree shrew (Tupaia belangeri) imported from Thailand (Kurz et al., 1986). DURV was isolated in the USA from an American coot (Fulica americana) with signs of neurological disease (Allison et al., 2011). KLAV has been isolated in the USA from a montane vole (Microtus montanus), northern red-backed voles (Clethrionomys rutilus) and tundra voles (Microtus oeconomus) (Johnson 1965, Allison et al., 2011) and neutralising antibodies to KLAV have been detected in deer of several species, American bison (Bison bison) and humans (Zarnke et al., 1983, Eldridge et al., 1987, Stansfield et al., 1988). TUPVSB was detected organ tissue of African marsh rats (Dasymys incomtus) collected in Kenya. WRpeTV1 was detected in Pearson’s horseshoe bats (Rhinolophus pearsonii), Wenzhou Myotis laniger tupavirus 1 (WMlaTV1; species Tupavirus laniger) was detected in Chinese water myotis bats (Myotis laniger), BtTVBS1 was detected in lesser brown horseshoe bats (Rhinolophus stheno) and BtTVBS2 was detected in Stoliczka's trident bats (Aselliscus stoliczkanus). All were were detected by metagenomic sequencing of bats collected in Yunnan Province, China.
Antigenicity
KLAV cross-reacts in indirect immunofluorescence tests with TUPV (Calisher et al., 1989). However, immune serum to a wide range of other animal rhabdoviruses failed to cross-react with KLAV in complement-fixation (CF) tests and immune serum to KLAV failed to react in CF tests with DURV (Tesh et al., 1983, Allison et al., 2011).
Species demarcation criteria
Viruses assigned to different species within the genus Tupavirus have several of the following characteristics: A) minimum amino acid sequence divergence of 5% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) can be distinguished in serological tests; and F) occupy different ecological niches as evidenced by differences in hosts and/or arthropod vectors.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| duck rhabdovirus | MH453812* | DKRV |
| shelduck rhabdovirus | MH453880* | SDRV |
| Wufeng shrew tupavirus 1 | OQ715715* | WfSTV1 |
| Wufeng bat tupavirus 2 | OQ715690 | WfBTV2 |
Virus names and virus abbreviations are not official ICTV designations.
* Coding region sequence incomplete

