Subfamily: Betarhabdovirinae
Genus: Dichorhavirus
Distinguishing features
Viruses assigned to the genus Dichorhavirus form a monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Dichorhaviruses produce short bacilliform virions that lack a clearly distinguishable lipid envelope and have bi-segmented genomes. They share several characteristics with alphanucleorhabdoviruses, betanucleorhabdoviruses and gammanucleorhabdoviruses, such as nuclear viroplasm, cytopathological effects in the nucleus, composition of structural proteins and gene order, sequence similarity and transcription mechanism. They are transmitted by false spider mites (genus Brevipalpus) in which they appear to replicate.
Virion
Morphology
Dichorhaviruses produce short, bacilliform virions, 40 × 100–110 nm in size. Virions appear not to be enveloped but may be found in association with host membranes (Kitajima et al., 2003, Kondo et al., 2009).
Nucleic acid
The negative-sense single-stranded RNA genome is bi-segmented. RNA 1 (6.4–6.7 kb) contains five genes and RNA 2 (6.0 kb) contains a gene coding for L (Kondo et al., 2006, Ramalho et al., 2014). The termini of each segment are complementary and all genes are separated by conserved intergenic regions. Anti-genomic (positive-sense) RNAs are also present as a minor component (10–20 %) in purified orchid fleck virus (OFV; species Dichorhavirus orchidaceae) virions (Kondo et al., 2009).
Proteins
Dichorhaviruses express the five canonical rhabdovirus structural proteins, nucleocapsid protein (N), phosphoprotein (P), matrix protein (M) and glycoprotein (G) and RNA polymerase (L). N, G and L have moderate to high sequence similarity with the cognate proteins of alphanucleorhabdoviruses, betanucleorhabdoviruses and gammanucleorhabdoviruses. The P3 protein is a putative movement protein (Kondo et al., 2006, Ramalho et al., 2014, Leastro et al., 2020). There are no reports of accessory genes in viruses of this genus. In OFV, the masses of the proteins are 49 kDa (N), 26 kDa (P), 38 kDa (P3), 20 kDa (M), 61 kDa (G) and 212 kDa (L).
Genome organisation and replication
The genome organisation of viruses in the genus Dichorhavirus resembles that of plant-infecting rhabdoviruses in the genera Cytorhabdovirus, Alphanucleorhabdovirus, Betanucleorhabdovirus, Deltanucleorhabdovirus and Gammanucleorhabdovirus, except that the RNA genome occurs as two segments: RNA 1 contains genes encoding five proteins in the order 3′-N-P-P3-M-G-5′ and RNA 2 codes for L (Figure 1 Dichorhavirus). Replication and transcription mechanisms closely resemble those reported for alphanucleorhabdoviruses, betanucleorhabdoviruses and gammanucleorhabdoviruses.
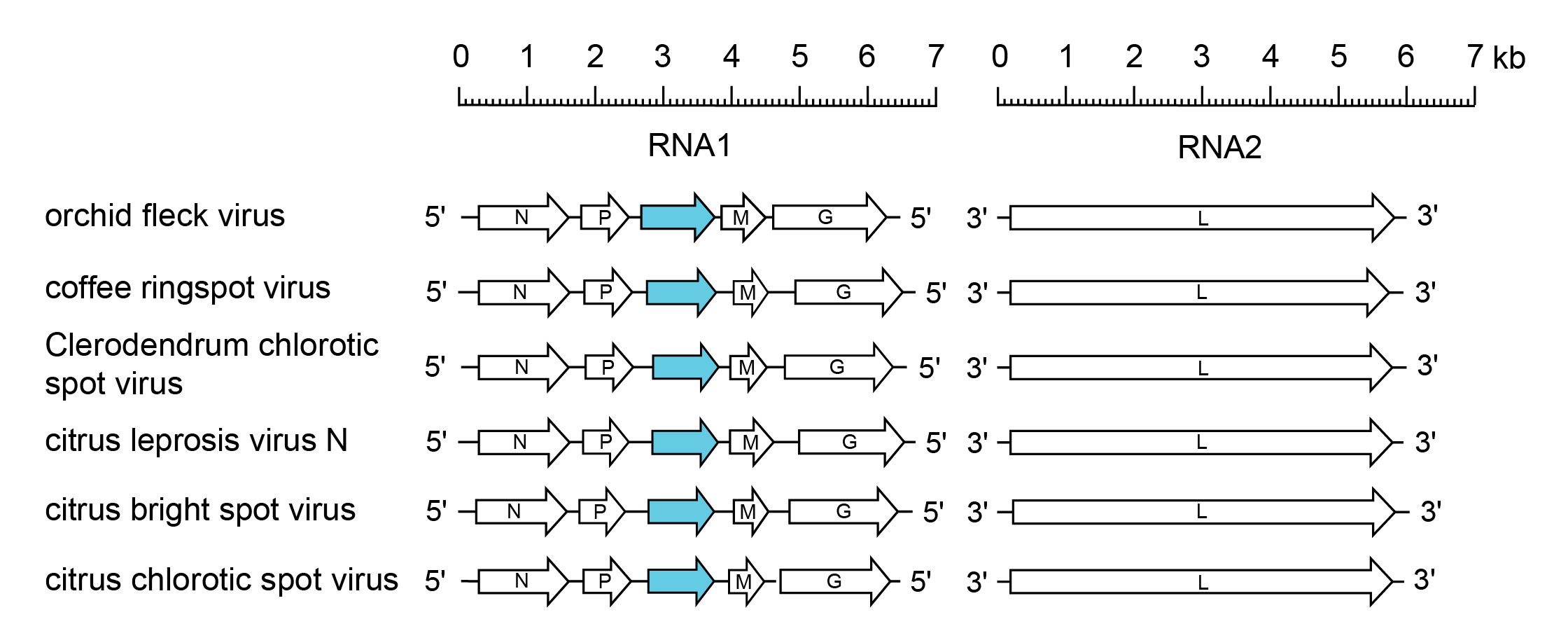 |
| Figure 1 Dichorhavirus. Schematic representation of dichorhavirus genome segments (RNA1 and RNA2) shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. ORF3, encoding the putative viral cell-to-cell movement protein gene, is highlighted (blue). |
Biology
Dichorhaviruses are transmitted by false spider mites of the genus Brevipalpus (Acari: Tenuipalpidae) in which they also probably replicate (Chagas et al., 2003, Kitajima et al., 2003, Kondo et al., 2003, Kitajima and Alberti 2014, Roy et al., 2015). Dichorhavirus infect monocots and dicots, leading to the appearance of local chlorotic and/or chlorotic spots on leaves and fruits (and/or systemic symptoms in some cases under high temperatures). They induce typical cytopathic effects in the nuclei of infected cells (Chang et al., 1976, Kitajima et al., 2001). While OFV infects orchids worldwide, other hosts such as citrus and ornamental plants (lilyturf, green cordyline and common hollyhock) have been reported in few locations (Peng et al., 2013, Mei et al., 2016, Dietzgen et al., 2018, Cook et al., 2019). Three dichorhaviruses, citrus leprosis virus N (CiLV-N; species Dichorhavirus leprosis), citrus bright spot virus (CiBSV; species Dichorhavirus australis) and citrus chlorotic spot virus (CiCSV; species Dichorhavirus citri), infect citrus in Brazil (Ramos-Gonzalez et al., 2017, Chabi-Jesus et al., 2018). Whereas CiLV-N and CiBSV seem to infect only sweet oranges (Chabi-Jesus et al., 2023), CiCSV has a broader host range which includes beach hibiscus and Agave desmettiana (Chabi-Jesus et al., 2019). Clerodendrum chlorotic spot virus (ClCSV; species Dichorhavirus clerodendri), also characterised in Brazil, infects the ornamental Clerodendrum sp. and a large number of host species (Kitajima et al., 2008, Ramos-González et al., 2018). Coffee ringspot virus (CoRSV; species Dichorhavirus coffeae) has been reported to infect coffee in Brazil where it seems to be endemic. It has also been reported in Costa Rica but has not been detected in that country since the first report. CoRSV genetic variability and epidemiology has been studied (Ramalho et al., 2016). CoRSV, CiLV-N, CiCSV, CiBSV and ClCSV are mite-transmitted to the model plant Arabidopsis thaliana, where chlorotic and necrotic spots are observed (Arena et al., 2017, Chabi-Jesus et al., 2023).
Species demarcation criteria
Viruses assigned to different species within the genus Dichorhavirus have at least two of the following characteristics: A) minimum nucleotide sequence divergence of 20% in L genes; B) minimum nucleotide sequence divergence of 20% in RNA1; C) can be distinguished in serological tests; and D) occupy different ecological niches as evidenced by differences in plant hosts and/or arthropod vectors.

