Subfamily: Betarhabdovirinae
Genus: Gammanucleorhabdovirus
Distinguishing features
Historically, the genera Nucleorhabdovirus and Cytorhabdovirus were established based on the sites of virus replication and morphogenesis, with nucleorhabdoviruses replicating and maturing in the nuclei of infected cells. Recently, a reclassification became necessary as phylogenetic analyses of new plant rhabdovirus genomes consistently showed that nucleorhabdoviruses did not form a monophyletic clade upon analysis of complete L protein sequence alignments (Kuhn et al., 2020).
Maize fine streak virus (MFSV; species Gammanucleorhabdovirus maydis) is the sole member of the genus Gammanucleorhabdovirus. Although MFSV is phylogenetically closest to the alphanucleorhabdoviruses, nucleotide and deduced amino acid sequence comparisons between them, as well as between MFSV and other plant rhabdoviruses, are extremely low. Additionally, phylogenetic analyses of L protein sequences indicate that bootstrap support for MFSV grouping with members of the genus Alphanucleorhabdovirus is very weak. Therefore, the genus Gammanucleorhabdovirus was created.
Virion
Morphology
Enveloped virions are bacilliform, 231 × 71 nm in size (Redinbaugh et al., 2002).
Nucleic acid
The negative-sense, single-stranded RNA genome of 13.8 kb is unsegmented. Seven mRNAs, one for each of the encoded proteins, have been identified in infected plants (Tsai et al., 2005).
Proteins
Purified preparations of MFSV contain three abundant proteins corresponding to the viral G, N and M proteins. Using in planta subcellular localization studies of overexpressed ORF fusions to autofluorescent proteins showed that N, P4 and M proteins accumulate in the nucleus, whereas P protein was spread throughout the cell and P3 accumulated in punctate loci in the cytoplasm. N and P proteins interacted and co-localized to the nucleolus (Tsai et al., 2005).
Lipids
The lipoprotein envelope is derived from the host plant or insect vector (Jackson et al., 2005a). Lipid composition is unknown.
Genome organisation and replication
The MFSV genome (13.8 kb) has the gene order 3′-N-P-P3-P4-M-G-L-5′- similar to that of the betanucleorhabdovirus Sonchus yellow net virus (SYNV; species Betanucleorhabdovirus retesonchi), but with an additional ORF of unknown function between the P gene and the M gene. The proteins encoded in ORFs P3 and P4 display no homology with those of other rhabdoviruses, including the P3 movement proteins of other plant rhabdoviruses (Tsai et al., 2005) (Figure 1.Gammanucleorhabdovirus).
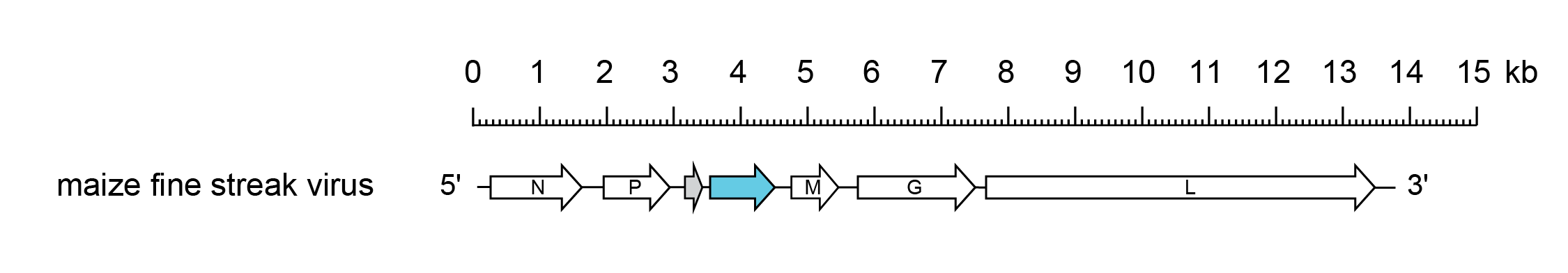 |
| Figure 1.Gammanucleorhabdovirus. Schematic representation of the maize fine streak virus genome shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. Other ORFs encode a putative movement protein (blue) or and accessory protein of unknown function (grey). |
MSFV replicates in the nuclei of plant cells, which become greatly enlarged and develop large granular nuclear inclusions that are thought to be sites of virus replication (Jackson et al., 2005a).
Biology
MFSV was originally identified in maize, but its host range extends to other gramineous plants. It can be experimentally transmitted by vascular puncture inoculation, but not by mechanical rub-inoculation of leaves. MFSV is transmitted in a persistent manner by a leafhopper (Graminella nigrifrons Forbes, 1885) in which it replicates. MFSV can be differentiated from other maize rhabdoviruses by serology using western blotting (Redinbaugh et al., 2002).
Species demarcation criteria
Only one virus, MFSV has been assigned to one species within the genus Gammanucleorhabdovirus. No species demarcation criteria have been developed thus far.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| cereal chlorotic mottle virus | MW731536 | CCMoV |
Virus names and virus abbreviations are not official ICTV designations.

