Subfamily: Alpharhabdovirinae
Genus: Sprivivirus
Distinguishing features
Viruses assigned to the genus Sprivivirus belong to one of the five rhabdovirus genera that infect finfish, the other genera being Perhabdovirus, Siniperhavirus, Scophrhavirus and Novirhabdovirus. Spriviviruses form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences and are most closely related to perhabdoviruses and vesiculoviruses. Viruses assigned to the genus have been isolated predominantly from cypriniform fish (order Cypriniformes).
Virion
Morphology
Spring viraemia of carp virus (SVCV; species Sprivivirus cyprinus) virions exhibit bullet-shaped morphology, and measure 120–180 nm in length and 60–90 nm in diameter (Ahne et al., 2002).
Nucleic acid
The sprivivirus genome consists of a single molecule of negative-sense, single-stranded RNA of approximately 11 kb.
Proteins
N, P, M, G and L share significant sequence homology and/or structural characteristics with the homologous proteins of other rhabdoviruses. SVCV G is an immune-protective antigenic protein; DNA vaccination using the SVCV G gene provides 48–80% relative protection (Kanellos et al., 2006, Emmenegger and Kurath 2008).
Carbohydrates
SVCV G is glycosylated and contains five possible sites for N-linked glycosylation (Zhang et al., 2009).
Genome organisation and replication
The sprivivirus genome contains five genes in the order 3′-N-P-M-G-L-5′ encoding a nucleoprotein, polymerase-associated protein, matrix protein, glycoprotein and RNA-directed RNA polymerase, respectively (Figure 1 Sprivivirus). The genome contains a leader region of approximately 59 nt preceding the transcription start of the N gene, and a trailer of about 19 nt following the transcription termination of the L gene. The putative transcriptional initiation and termination/polyadenylation signals are conserved for all genes; 3′-UUGUC and 3′-AURC(U)7 respectively, and the non-transcribed intergenic regions are 3′-GA, except the G-L intergenic region which is 3′-GAUA. There is inverse complementarity between the 3′-leader and 5′-trailer sequences. The putative transcript encoding the SVCV G gene varies in length by up to 85 nt due to variation in the 3′-untranslated region.
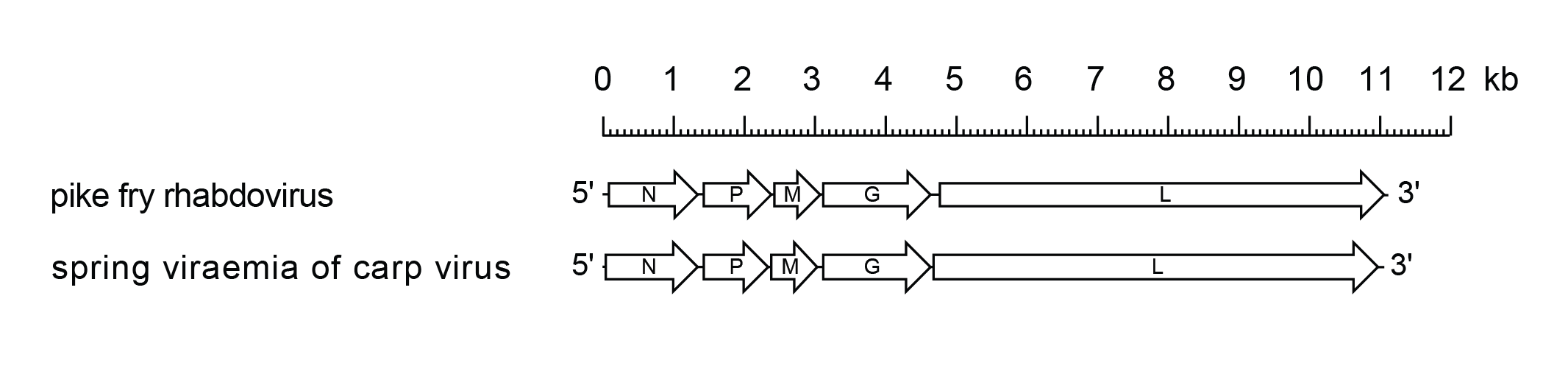 |
| Figure 1 Sprivivirus. Schematic representation of sprivivirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. |
Biology
Naturally occurring SVCV infections have been recorded predominantly cyprinid fish including common carp (Cyprinus carpio carpio) and koi carp (Cyprinus carpio koi), bighead carp (Aristichthys nobilis), crucian carp (Carassius carassius), grass carp (Ctenopharyngodon idella), silver carp (Hypophthalmichthys molitrix), goldfish (Carassius auratus), orfe (Leuciscus idus), tench (Tinca tinca), roach (Rutilus rutilus) bream (Abramis brama) and emerald shiner (Notropis atherinoides) (Basic et al., 2009, Cipriano et al., 2011). Non-cyprinid hosts include rainbow trout (Oncorhynchus mykiss), sheatfish (Silurus glanis), pike (Esox lucius), Siberian sturgeon (Acipenser baerii), largemouth bass (Micropterus salmoides) and bluegill sunfish (Lepomis macrochirus) (Cipriano et al., 2011, Vicenova et al., 2011, Phelps et al., 2012). Pike fry rhabdovirus (PFRV; species Sprivivirus esox) has been reported in pike, sheatfish and grass carp.
The replication temperature range of SVCV is lower than those of mammalian rhabdoviruses, reflecting the aquatic poikilothermic nature of the host species, and the viruses are typically isolated in cultured fish cell lines at 15–25 °C. The disease patterns are influenced by water temperature, age and condition of the fish, population density and stress factors. The immune status of the fish is also an important factor with both innate and adaptive immunity having important roles. Clinical disease is usually observed at water temperature between 5–18 °C and is most severe at temperatures below 10 °C when it is believed the host immune response is suppressed or delayed.
SVCV transmission occurs horizontally (Fijan 1988). Transmission can involve ectoparasite vectors such as carp lice (Argulus foliaceus) and leeches (Pisicola geometra) but direct waterborne transmission is also effective. The virus has been isolated from ovarian fluid (Bekesi and Csontos 1985) but vertical transmission has not been demonstrated. SVCV appears to enter fish via the gills (Dixon 2008) and then rapidly spreads to the kidney, liver, heart, spleen and alimentary tract. During disease outbreaks, high titres of virus are detected in the liver and kidneys of infected fish, whereas lower titres are found in the gills, spleen and brain (Fijan et al., 1971). Shedding of SVCV by survivors is probably the primary mechanism of transmission but not much is known about the persistence of the virus in infected fish, the duration of viral shedding or amounts of virus shed. The virus may also be shed following a stressful event, particularly from fish in poor condition in the spring following a harsh winter (Fijan 1988) and during spawning.
Many SVCV strains have been identified. Based on nucleotide sequence analysis of the G gene, SVCV isolates are further classified into four genogroups: Ia, Ib, Ic and Id (Stone et al., 2003, Miller et al., 2007, Warg et al., 2007, Zhang et al., 2009). The genetic clustering of SVCV isolates into genogroups is closely associated with geographical location, suggesting that the virus has evolved independently in different geographical regions (Stone et al., 2013). Genogroup Ia contains isolates from Asia, the UK and North America; Ib and Ic contain isolates from Eastern Europe; and Id contains isolates from the UK and some other European countries (Stone et al., 2003, Miller et al., 2007, Zhang et al., 2009, Padhi and Verghese 2012). Molecular epidemiology data provides evidence for international transport of SVCV in association with aquaculture-related movement of fish (Miller et al., 2007).
Antigenicity
SVCV occurs as a single recognised serotype. SVCV antibodies cross-react to various degrees with viruses assigned to the species Sprivivirus esox (Jorgensen et al., 1989, Dixon and Longshaw 2005). SVCV and PFRV G, N and M share common antigenic determinants but the viruses can be differentiated by neutralisation assays.
Species demarcation criteria
Viruses assigned to different species within the genus Sprivivirus have the following characteristics: A) minimum nucleotide sequence divergence of 30% in G genes; and B) can be clearly distinguished antigenically in serological tests.
SVCV and PFRV are distinguished serologically based on the lack of cross-neutralisation with polyclonal antisera, and virus strains of SVCV are neutralised by a single polyclonal antiserum (Ahne et al., 1988, Jorgensen et al., 1989). PFRV strain F4 shares 65–66% nt identity with SVCV strains based on partial G gene sequences (Stone et al., 2003), while SVCV viruses share 82.7–100% nt identity in the same genomic region. As occurs with many fish viruses, there is a broad and overlapping host range for viruses assigned to the genus Sprivivirus.
Discrimination between viruses assigned to the species Sprivivirus esox (PFRV, tench rhabdovirus [TRV] and grass carp rhabdovirus [GCRV]) using serum neutralisation is more difficult and is heavily dependent on the quality of the antiserum; therefore, distinction between viruses within the species is based on nucleotide sequence divergence of partial G gene sequences. TRV isolates share > 93.7% identity and GCRV isolates share > 98.5% identity. However, PFRV isolates share 82.0–83.5% identity with TRV isolates; PFRV isolates share 75–75.5% identity with GCRV isolates; and the GCRV and TRV isolates share between 71.5–72.5% identity. TRV, GCRV and PFRV appear to represent separate lineages within a single species and could be considered strains of the same virus.

