Subfamily: Alpharhabdovirinae
Genus: Sripuvirus
Distinguishing features
Viruses assigned to the genus Sripuvirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Viruses assigned to the genus have been isolated from either lizards or phlebotomine sandflies. Sripuviruses share the common features of: i) a consecutive ORF (Mx) in the M gene for which the initiation codon overlaps the termination codon of the M ORF; and ii) an ORF (Gx) in an alternative reading frame near the start of the G gene.
Virion
Morphology
In ultra-thin sections of infected cells, virions display long bullet-shaped or bacilliform morphology (i.e., with two rounded ends). For Niakha virus (NIAV; species Sripuvirus niakha), bacilliform virions (180–200 nm × 45 nm) have been reported in mammalian (BHK-21) cell cultures; in insect (Phlebotomus papatasi) cell cultures virions were observed to be up to 350 nm in length but these may represent longitudinally fused particles (Vasilakis et al., 2013). For Chaco virus (CHOV; species Sripuvirus chaco), long bullet-shaped particles (mean length = 202 nm) have been observed in infected cells (Monath et al., 1979).
Nucleic acid
Sripuvirus genomes consist of a single molecule of negative-sense, single-stranded RNA and range from approximately 11.0–11.5 kb (Walker et al., 2015).
Proteins
N, P, M, G and L share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses. Other proteins are encoded in the genome but have not yet been identified in infected cells. The Mx ORFs present in all sripuvirus genomes encode small basic proteins of 79 to 81 amino acids (9.4–9.7 kDa). The Gx ORFs also present in all sripuvirus genomes encode predicted small membrane-spanning proteins of 94 to 99 amino acids (10.4–11.1 kDa). The U1 ORFs in CHOV and Sena Madureira virus (SMV; species Sripuvirus madureira) genomes each encode a small highly acidic protein of 115 amino acids (13.6–13.7 kDa). The Almpiwar virus (ALMV; species Sripuvirus almpiwar) and Sripur virus (SRIV; species Sripuvirus sripur) Px ORFs encode highly basic proteins of 126 and 183 amino acids, respectively (15.4 kDa and 21.9 kDa) (Vasilakis et al., 2013, Walker et al., 2015).
Genome organisation and replication
Sripuvirus genomes include five genes (N, P, M, G and L) encoding the structural proteins and multiple additional long ORFs (Figure 1 Sripuvirus). The genomes of all sripuviruses feature a consecutive ORF (Mx) in the M gene for which the initiation codon overlaps the termination codon of the M ORF. “Termination upstream ribosome-binding site” (TURBS)-like sequence motifs typically occur upstream of the overlapping stop-start codons, suggesting the mechanism for expression. Sripuvirus genomes also feature an alternative ORF (Gx) near the start of the G gene. The Gx proteins are likely to be expressed by “leaky” ribosomal scanning. Two of the viruses (CHOV, SMV) have an additional gene (U1) between the N gene and P gene; two other sripuviruses (SRIV and ALMV) have an alternative long ORF (Px) near the start of the P gene (Vasilakis et al., 2013, McAllister et al., 2014, Walker et al., 2015).
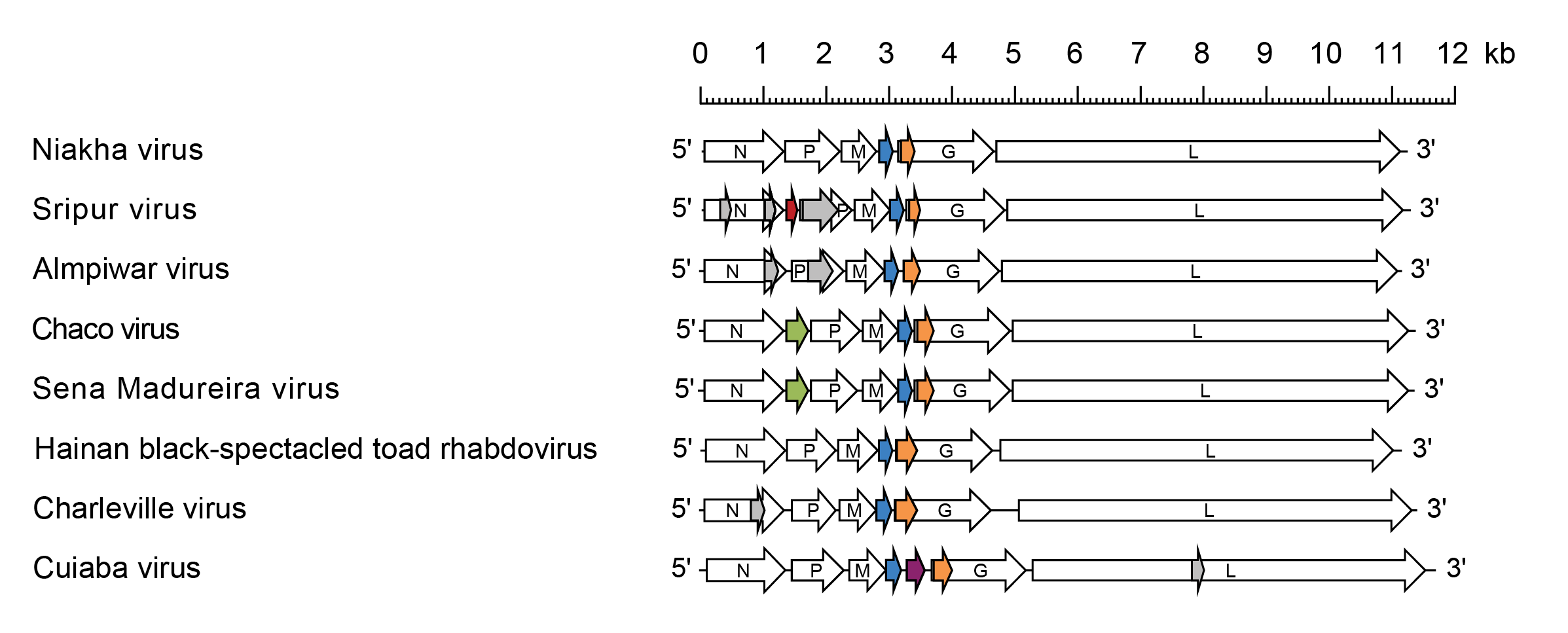 |
| Figure 1 Sripuvirus. Schematic representation of sripuvirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. ORF Mx (blue), ORF Gx (orange) and ORF U1 (green) encoding homologous sets of accessory proteins are highlighted. ORF U1 of Sripur virus (red brown) and ORF U1 of Cuiaba virus (purple) occur in independent transcriptional units and encode unique proteins. Alternative ORFs (≥180 nt) also occur in the N, P and L genes of some viruses (grey) but it is not known if they are expressed. |
Biology
Viruses assigned to the genus infect reptiles and appear to be transmitted by phlebotomine sandflies (Phlebotomus spp. or Seregentomyia spp.) (Causey et al., 1966, Doherty et al., 1970, Fontenille et al., 1994). A broader range of vertebrate hosts is possible as each replicates in mammalian cell cultures. The known distribution of members of the genus includes West Africa, South Asia, South America and Australia.
Antigenicity
Antigenic cross-reactions (complement-fixation test) have been reported between some members of the genus and these viruses (CHOV and SMV) have been assigned to the Timbo serogroup (Tesh et al., 1983, Calisher et al., 1989). Timbo virus (see below) is a related, unclassified rhabdovirus and also a likely member of the genus.
Species demarcation criteria
Viruses assigned to different species within the genus Sripuvirus have several of the following characteristics: A) minimum amino acid sequence divergence of 5% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) can be distinguished in virus neutralisation tests; and F) occupy different ecological niches as evidenced by differences in hosts and/or arthropod vectors.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Humpty Doo virus | AY854643* | HDOOV |
| Timbo virus | JN882646* | TIMV |
Virus names and virus abbreviations are not official ICTV designations.
* Coding region sequence incomplete

