Subfamily: Deltarhabdovirinae
Genus: Alphacrustrhavirus
Distinguishing features
Viruses assigned to the genus Alphacrustrhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Members of the genus have been detected in marine crustaceans (Crustacea).
Virion
Morphology
Viruses assigned to the genus have not yet been visualized by electron microscopy.
Nucleic acid
Alphacrustrhavirus genomes consist of a single molecule of negative-sense, single-stranded RNA of approximately 11.6–12.0 kb (Shi et al., 2016).
Proteins
Alphacrustrhavirus N, P, M, G and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses. Alphacrustrhavirus G proteins are class I transmembrane glycoproteins. Alignment with the G protein of vesicular stomatitis Indiana virus indicates likely conservation of 8 of the 12 conserved cysteine residues that are typical of animal rhabdovirus G proteins (Walker and Kongsuwan 1999, Roche et al., 2006).
Genome organisation and replication
Alphacrustrhavirus genomes include the five genes (N, P, M, G and L) encoding the structural proteins (Figure 1 Alphacrustrhavirus). In Wenling crustacean virus 11 (WlCV11; species Alphacrustrhavirus zhejiang), the P gene contains an alternative open reading frame (ORF) of 297 nt encoding a highly basic 11.9 kDa protein (Px). The Px ORF commences near the start of the P ORF but it is not known if the protein is expressed.
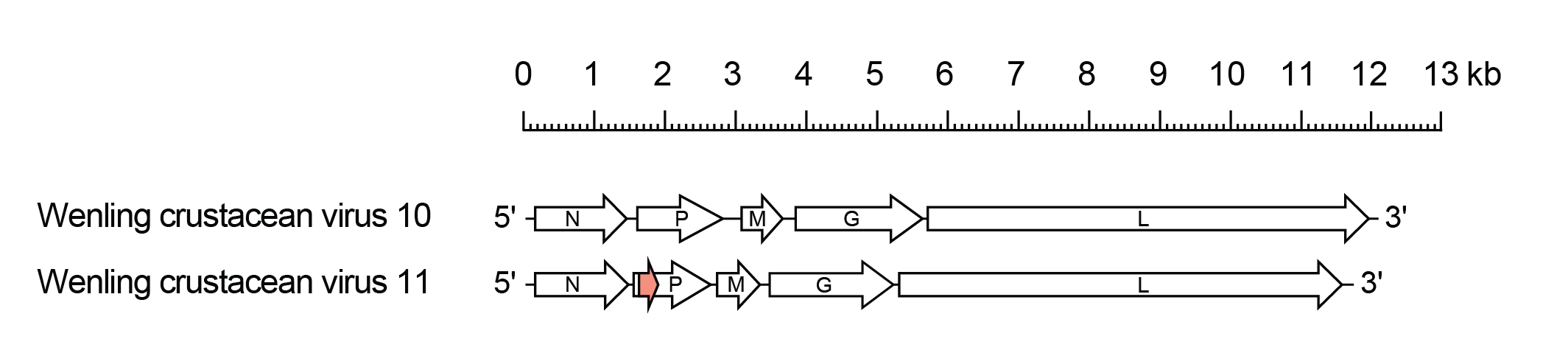 |
| Figure 1 Alphacrustrhavirus. Schematic representation of alphacrustrhavirus genomes shown in reverse (positive-sense) polarity. The five long open reading frames (ORFs) in the N, P, M, G and L genes are shown (open arrows). In Wenling crustacean virus 11, there is an alternative ORF (Px) in the P gene (orange). |
Biology
WlCV11 and Wenling crustacean virus 10 (WlCV10; species Alphacrustrhavirus wenling) were each discovered in a pool of marine crustaceans (multiple families) collected in Zhezhang Province, China, in 2014 (Shi et al., 2016). No isolates are currently available for either of these viruses.
Species demarcation criteria
Viruses assigned to different species within the genus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) can be distinguished in virus neutralisation tests; and F) occupy different ecological niches as evidenced by differences in invertebrate hosts.

