Subfamily: Alpharhabdovirinae
Genus: Lostrhavirus
Distinguishing features
Viruses assigned to the genus Lostrhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Lostrhaviruses have been detected in hard ticks (Ixodidae). Lone star tick rhabdovirus (LSTRV; species Lostrhavirus lonestar) was detected in a tick collected from a patient with a rash illness.
Virion
Morphology
The virion morphology is unknown.
Nucleic acid
The lostrhavirus genome consists of a single molecule of negative-sense, single-stranded RNA of approximately 11.5 kb.
Proteins
The lostrhavirus N, P, M, G and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses.
Genome organisation and replication
The lostrhavirus genome includes only the five genes (N, P, M, G and L) encoding the structural protein (Figure 1 Lostrhavirus). There are alternative ORFs (≥180 nt) in the N and P genes but it is not known if they are functional.
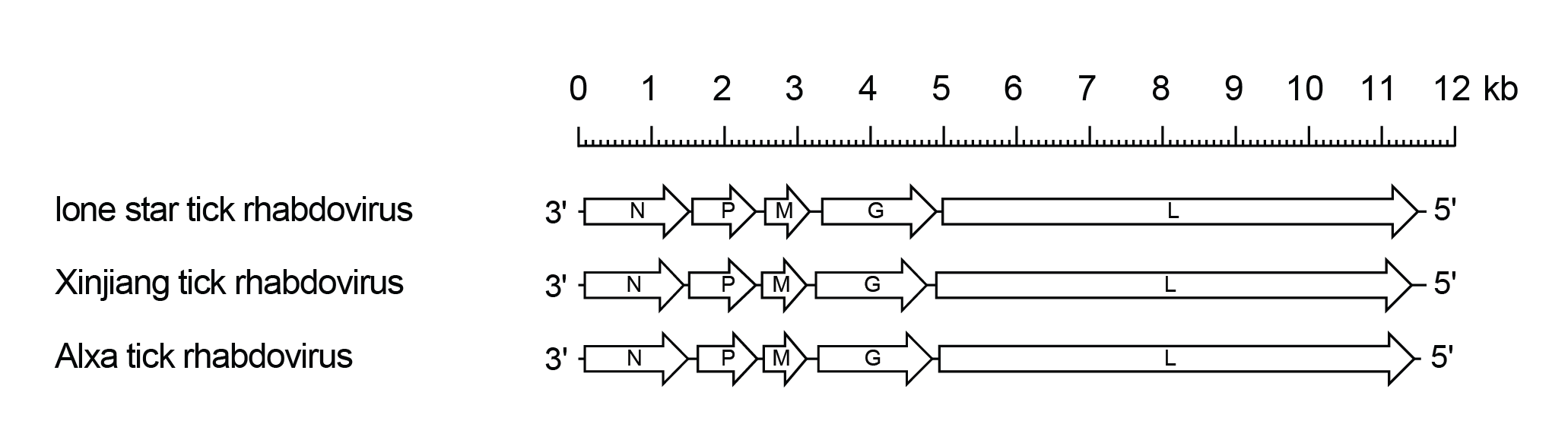 |
| Figure 1 Lostrhavirus. Schematic representation of lostrhavirus genomes shown in reverse (positive-sense) polarity. The genomes contain five long open reading frames (ORFs) in the N, P, M, G and L genes (open arrows). |
Biology
Lostrhaviruses have been detected in hard ticks (Ixodidae) of different genera. Xinjiang tick rhabdovirus (XjTRV; species Lostrhavirus hyalomma) was isolated from Asiatic hyalomma ticks (Hyalomma asiaticum) in China. Alxa tick rhabdovirus (ATRV; species Lostrhavirus alxa) was detected by metagenomic sequencing of hard ticks (Hyalomma sp.) collected from herbivore livestock in Xinjiang and Inner Mongolia Autonomous Regions, China. LSTRV was isolated from lone star ticks (Amblyomma americanum) collected from a patient in the USA with a rash illness.
Species demarcation criteria
Viruses assigned to different species within the genus Lostrhavirus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in the N proteins; B) minimum amino acid sequence divergence of 10% in the the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) they can be distinguished in virus neutralisation tests; and F) they occupy different ecological niches as evidenced by differences in vertebrate hosts and or arthropod vectors.

