Subfamily: Alpharhabdovirinae
Genus: Sawgrhavirus
Distinguishing features
Viruses assigned to the genus Sawgrhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Viruses assigned to the genus have been isolated from hard ticks taken from small mammals and marsupials. They are most closely related to rhabdoviruses in the genus Mousrhavirus, the sole member of which was isolated from mosquitoes.
Virion
Morphology
Bullet-shaped particles of Sawgrass virus (SAWV; species Sawgrhavirus sawgrass) and New Minto virus (NMV; species Sawgrhavirus minto), 220 nm in length, have been observed budding from neuronal plasma membranes and accumulating within extracellular spaces in the brains of experimentally infected newborn mice (Ritter et al., 1978).
Nucleic acid
Sawgrhavirus genomes consist of a single molecule of negative-sense, single-stranded RNA of approximately 12.2 kb (Walker et al., 2015).
Proteins
Sawgrhavirus N, P, M, G and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses. Putative proteins Gx and Gy encoded in alternative small ORFs in the G genes of some sawgrharviruses have no distinguishing structural characteristics.
Genome organisation and replication
Sawgrhavirus genomes include five genes (N, P, M, G and L) encoding the structural protein (Walker et al., 2015) (Figure 1 Sawgrhavirus). Two alternative ORFs (Gx and Gy) in the G genes of Connecticut virus (CNTV; species Sawgrhavirus connecticut) and Long Island tick rhabdovirus (LITRV; species Sawgrhavirus longisland) share identifiable sequence similarity. Alternative ORFs occur in some genes of these and other sawgrhaviruses but are not conserved. It is not known if any of these alternative ORFs are expressed.
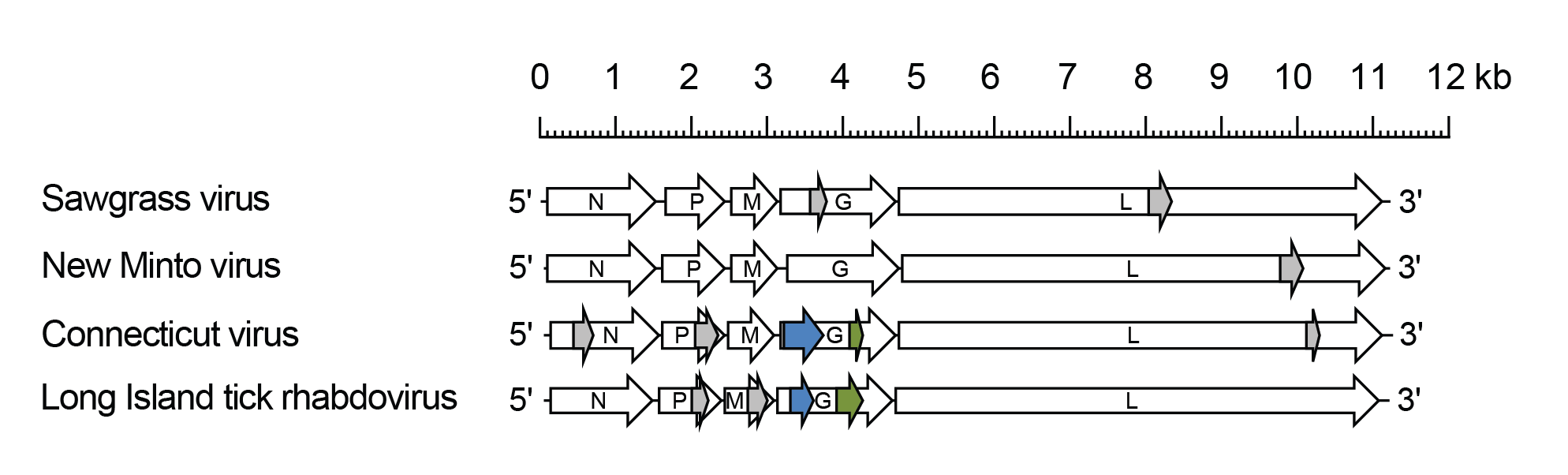 |
| Figure 1 Sawgrhavirus. Schematic representation of sawgrhavirus genomes shown in reverse (positive-sense) polarity. Each genome contains long open reading frames (ORFs) in the N, P, M, G and L genes (open arrows). Alternative ORFs Gx (blue) and Gy (green) in the G genes of Connecticut virus and Long Island tick rhabdovirus are shown. ORFs (≥180 nt) with no obvious homologues are also shown (grey). |
Biology
Sawgrhaviruses have been isolated from or detected in hard ticks (Acari: Iodidae) collected in the USA. SAWV was first isolated from Dermacentor variabilis ticks taken from an opossum captured in Florida, in 1964 (Sather et al., 1970). Eight other isolates of the virus were obtained from the same species of tick taken from opossums, racoons and rabbits, and from hard ticks of another species (Haemaphysalis leporispalustris) taken from rabbits captured in the same area in 1964 (Sather et al., 1970). A further 19 isolates were obtained from hard ticks of the same two species in 1968 and 1969 (Wellings et al., 1972). NMV was isolated on three occasions from Haemaphysalis leporispalustris ticks removed from snowshoe hares (Lepus americanus) captured in Alaska, in 1972 (Ritter et al., 1978). CNTV was isolated from Ixodes dentatus ticks taken from an eastern cottontail rabbit (Sylvilagus floridanus) captured in Connecticut, in 1978 (Main and Carey 1980). Neutralising antibody to CNTV has been detected in eastern cottontail rabbits (Main and Carey 1980). Long Island tick rhabdovirus (LITRV; strain LS1) was detected in Amblyomma americanum ticks collected in New York, in 2013 (Tokarz et al., 2014). CNTV, SAWV and NMV cross-react weakly in complement-fixation and neutralisation tests and have been described as the ‘Sawgrass serogroup’ of rhabdoviruses (Tesh et al., 1983).
Species demarcation criteria
Viruses assigned to different species within the genus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) they can be distinguished in virus neutralisation tests; and F) they occupy different ecological niches as evidenced by differences in vertebrate hosts and or arthropod vectors.

