Subfamily: Alpharhabdovirinae
Genus: Sunrhavirus
Distinguishing features
Viruses assigned to the genus Sunrhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Viruses assigned to the genus have been isolated from culicine mosquitoes, biting midges or birds in Central Africa, Egypt, North America or Australia.
Virion
Morphology
Bullet-shaped Kwatta virus (KWAV; species Sunrhavirus kwatta) virions have been observed in ultrathin sections of infected mouse brain and infected Vero cells (Karabatsos et al., 1973).
Nucleic acid
Sunrhavirus genomes consist of a single molecule of negative-sense, single-stranded RNA of approximately 10.1–12.5 kb (Ledermann et al., 2014, McAllister et al., 2014, Walker et al., 2015, Luo et al., 2021).
Proteins
Sunrhavirus N, P, M, G and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses. The KWAV, Oak Vale virus (OVV; species Sunrhavirus oakvale) and Dillard’s Draw virus (DDRV; species Sunrhavirus dillard) small hydrophobic proteins (SH) share identifiable sequence homology. The SH proteins of other sunrhaviruses are homologous in predicted structure and display identifiable sequence homology with the SH proteins of rhabdoviruses in the genus Tupavirus. However, they do not display obvious sequence homology with the KWAV, OVV or DDRV SH proteins.
Genome organisation and replication
Sunrhavirus genomes include five genes (N, P, M, G and L) encoding the structural proteins and an additional transcriptional unit located between the M and G genes (U1 gene) encoding a small hydrophobic protein (SH) (Figure 1 Sunrhavirus). Sunrhavirus P genes also feature an alternative ORF (Px) commencing near the start of the P ORF. Although there are significant size variations, pairwise alignments indicate that there is sequence identity between sunrhavirus Px proteins. Harrison Dam virus (HARDV; species Sunrhavirus harrison) and Boteke virus (BOTV; species Sunrhavirus boteke) also have an additional gene (U2) between the G and L genes, but the encoded proteins display no evident hsequence homology. Although these are complete and independent transcriptional units (i.e., bounded by conserved transcription initiation and transcription termination/polyadenylation sequences), the HARDV U2 gene encodes a protein of only 19 amino acids (2.38 kDa).
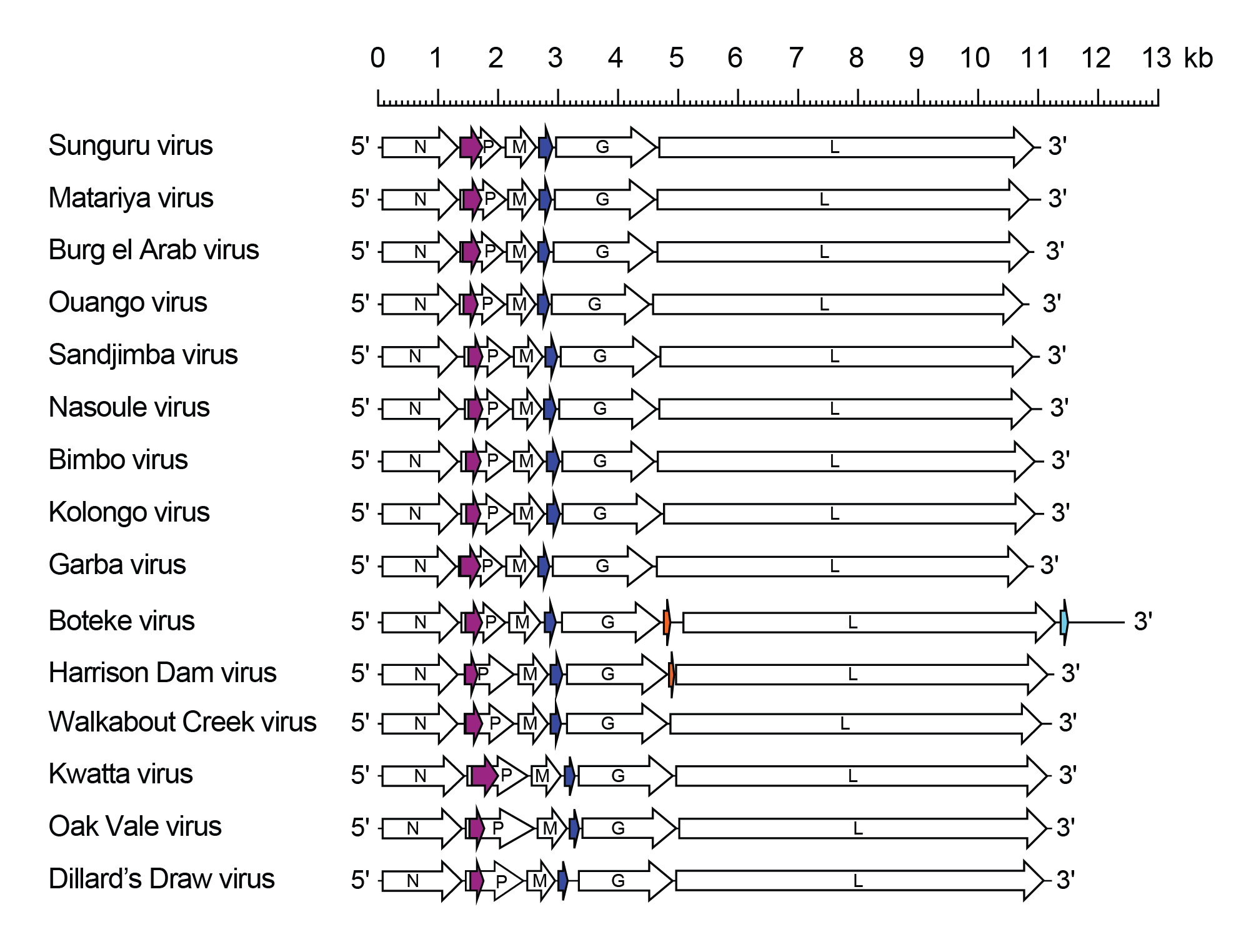 |
| Figure 1 Sunrhavirus. Schematic representation of ohlsrhavirus genomes shown in reverse (positive-sense) polarity. Each genome contains long open reading frames (ORFs) in the N, P, M, G and L genes (open arrows). Additional ORFs between the M and G genes (dark blue) occur in independent transcriptional units (genes) and encode small hydrophobic proteins. Alternative ORFs near the start of the P ORFs are shown (purple). In Poteke and Harrisin Dam viruses there is a small ORF (orange) within an additional transcriptional unit between the G and L genes. In Boteke virus there also is an additional ORF (light blue) in a small transcriptional unit in the long nucleotide sequence following the L gene; two other ORFs (≥180 nt) in this region are not bounded by transcriptional regulatory sequences and so cannot be expressed. |
Biology
Sunrhaviruses have been isolated from or detected in culicine mosquitoes (Diptera: Culicidae), biting midges (Ceratopogonidae) and birds collected in Central Africa, Egypt, North America or Australia. KWAV was isolated from Culex spp. mosquitoes collected in Surinam, in 1964 (De Haas et al., 1966). BOTV, Garba virus (GARV; species Sunrhavirus garba), Sandjimba virus (SJAV; species Sunrhavirus sandjimba), Nasoule virus (NASV; species Sunrhavirus nasoule), Bimbo virus (BBOV; species Sunrhavirus bimbo), Kolongo virus (KOLV; species Sunrhavirus kolongo), Ouango virus (OUAV; species Sunrhavirus ouango) were all isolated from birds of various species trapped in the Central African Republic from 1968 to 1973 (Karabatsos 1985, Luo et al., 2021). Sunguru virus (SUNV; species Sunrhavirus sunguru) was isolated from a domestic chicken in Uganda, in 2011 (Ledermann et al., 2014). Burg el Arab virus (BEAV; species Sunrhavirus alexandria) and Matariya virus (MTYV; species Sunrhavirus matariya) were isolated from birds trapped in Egypt in 1961 and 1962 (Luo et al., 2021). HARDV was isolated from Culex annulirostris mosquitoes collected in the Northern Territory, Australia, in 1975 (Standfast et al., 1984, McAllister et al., 2014). Trace levels of neutralizing antibodies to HARDV have been detected in horses and crocodiles (McAllister et al., 2014). Walkabout Creek virus (WACV; species Sunrhavirus walkabout) was isolated from biting midges (Culicoides austropalpalis) collected in Queensland, Australia, in 1981 (McAllister et al., 2014). OVV was isolated on multiple occasions from Culex edwardsi mosquitoes in Queensland, Australia, in 1981/82 (Cybinski and Muller 1990), and subsequently from Anopheles annulipes mosquitoes collected Western Australia, in 1993 (Quan et al., 2011). There is a report of neutralizing antibodies to OVV in feral pigs (Bourhy et al., 2008). DDRV was detected by metagenomic sequencing in mosquitoes (Culex tarsalis) collected in New Mexico, in 2015 (Reeves et al., 2018).
Species demarcation criteria
Viruses assigned to different species within the genus Sunrhavirus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organization as evidenced by numbers and locations of ORFs; E) they can be distinguished in virus neutralisation tests; and F) they occupy different ecological niches as evidenced by differences in vertebrate hosts and or arthropod vectors.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| BeAn 157575 virus | JX276982* | - |
| Navarro virus | JX277013* | NAVV |
Virus names and virus abbreviations are not official ICTV designations.
* Coding region sequence incomplete

