Subfamily: Alpharhabdovirinae
Genus: Perhabdovirus
Distinguishing features
Viruses assigned to the genus Perhabdovirus belong to one of the five genera for rhabdoviruses that infect finfish, the other genera being Siniperhavirus, Scophrhavirus, Sprivivirus and Novirhabdovirus. Perhabdoviruses form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. They are most closely related to siniperhaviruses, scophrhaviruses and cetarhaviruses (infecting cetaceans). Viruses assigned to the genus have been isolated from a wide range of fish but they predominantly cause disease in farmed perciform fishes (order Perciformes, perch-like) and eels (order Anguilliformes).
Virion
Morphology
Perch rhabdovirus (PRV; species Perhabdovirus perca) virions have a compact bullet-shaped morphology and measure 115–130 nm in length and 85–95 nm in diameter (Betts et al., 2003).
Nucleic acid
The genome consists of a single molecule of negative-sense, single-stranded RNA of approximately 11 kb.
Proteins
N, P, M, G and L share significant sequence homology with the homologous proteins of other rhabdoviruses.
Genome organisation and replication
The genomic RNA is approximately 11.4–11.8 kb with five genes in the order 3′-N-P-M-G-L-5′ encoding a nucleoprotein, polymerase-associated protein, matrix protein, glycoprotein and RNA-directed RNA polymerase, respectively (Figure 1 Perhabdovirus). The genome contains a leader region of approximately 63–100 nt preceding the transcription initiation of the N gene, and a trailer of 40–67 nt following the transcription termination of the L gene. The transcriptional initiation and the termination/polyadenylation signals are conserved for all genes, 3′-UUGUC and 3′-AURC(U)7, respectively, but the non-transcribed intergenic regions are variable. There is inverse complementarity between the 3′-leader and 5′-trailer sequences.
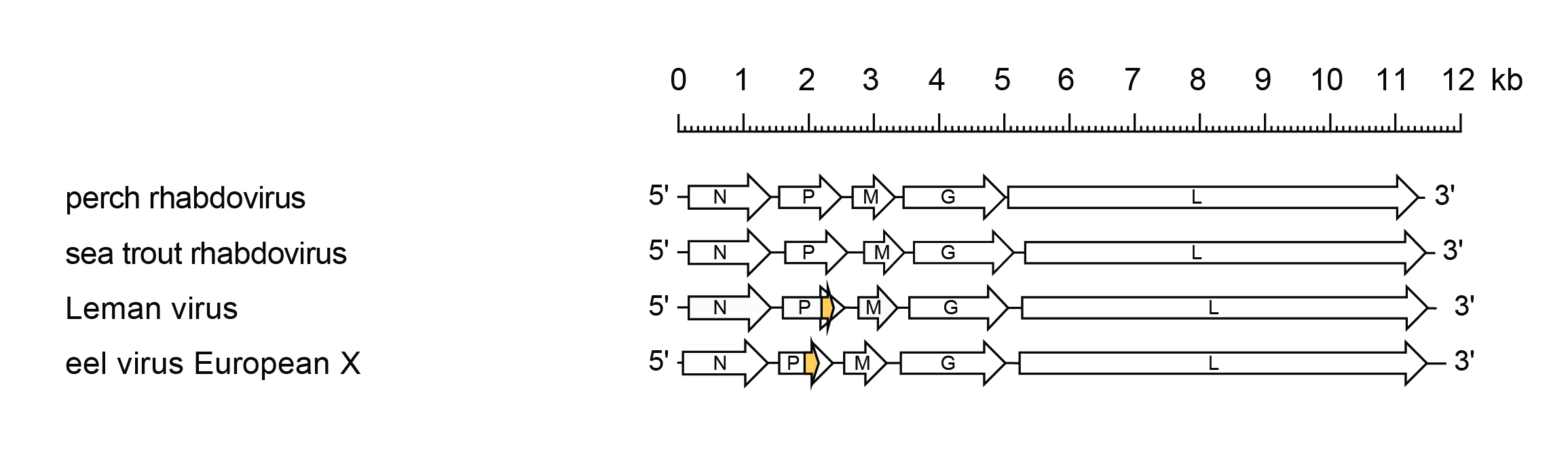 |
| Figure 1 Perhabdovirus. Schematic representation of perhabdovirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. A small alternative ORF occurs in the P gene of some perhabdoviruses (orange) but it is not known if these are expressed as functional proteins. |
Biology
PRV was first isolated from cultured perch (Perca fluviatilis) following a mortality event in France (Dorson et al., 1984) and subsequently from perch in Norway (Dannevig et al., 2001). Antigenically similar viruses have been recovered from diseased pike perch (Stizostedion lucioperca) (Nougayrède et al., 1992), grayling (Thymallus thymallus) and largemouth bass (Micropterus salmoides) in France (Betts et al., 2003) and from asymptomatic pike in Denmark (Jorgensen et al., 1989). These viruses are all assigned to the species Perhabdovirus perca. Leman virus was isolated from perch fry in France and has been assigned to the species Perhabdovirus leman (Pallandre et al., 2020). SSTV was discovered in brown trout (Salmo trutta m. lacustris) in Finland (Jorgensen et al., 1989, Koski et al., 1992, Björklund et al., 1994) and sea trout (Salmo trutta trutta) from Sweden (Johansson et al., 2002). More recently, viruses assigned to the species Perhabdovirus trutta have been isolated from perch, eel and brown trout in Ireland (Ruane et al., 2014). Serologically related viruses have been isolated from eels in Japan (Sano et al., 1977, Kobayashi and Miyazaki 1996). Eel virus American was isolated from American elvers (Anguilla rostrate) and eel virus European X was isolated from European eels (Anguilla anguilla); both are assigned to the species Perhabdovirus anguilla.
As for other fish rhabdoviruses, the replication temperature range for perhabdoviruses is lower than for mammalian rhabdoviruses, reflecting the aquatic poikilothermic nature of the hosts. Viruses are typically isolated on cultured fish cell lines at 15–25 °C. The disease patterns are influenced by water temperature, age and condition of the fish, population density and stress factors. The immune status of the fish is also an important factor with both innate and adaptive immunity having important roles. Clinical disease is usually observed at water temperature between 5–18 °C and is most severe at temperatures below 10 °C when it is believed the host immune response is suppressed or delayed.
Antigenicity
Antisera to the reference isolate of PRV efficiently neutralise PRV isolates from fish of a range of species (Dorson et al., 1984) but there is no cross-neutralisation between PRV and spring viraemia of carp virus (genus Sprivivirus) or viral haemorrhagic septicaemia virus (genus Novirhabdovirus) (Betts et al., 2003).
Species demarcation criteria
Viruses assigned to different species within the genus Perhabdovirus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in N proteins; B) minimum sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in G proteins; D) can be distinguished in virus neutralisation tests; and E) occupy different ecological niches as evidenced by differences in vertebrate hosts.

