Classification and genotype / subtype assignments of hepatitis C virus
Donald B. Smith, Jens Bukh, Carla Kuiken, A. Scott Muerhoff, Charles M. Rice, Jack T. Stapleton and Peter Simmonds
This web page maintains a regularly updated list of confirmed hepatitis C virus (HCV) genotype and subtype assignments (Table 1) and alignments (in FASTA and SSE formats) This maintained resource is designed to assist researchers investigating the diversity of HCV in designating appropriate assignments for new sequences and maintaining consistency in both the names designated and ensuring that guidelines for their assignment (originally proposed in the published consensus paper (1) and updated recently (2)) are appropriately followed. The ICTV Online (10th) Report describing the taxonomic position of HCV is available at https://ictv.global/report/flaviviridae (3).Although the 2005 proposal was supported by by the establishment of curated databases that organised HCV sequences as they became available (Los Alamos HCV Sequence Database, (4) and euHCVdb (5)), neither of these are now actively maintained. This web resource is designed to maintain this function for HCV researchers.
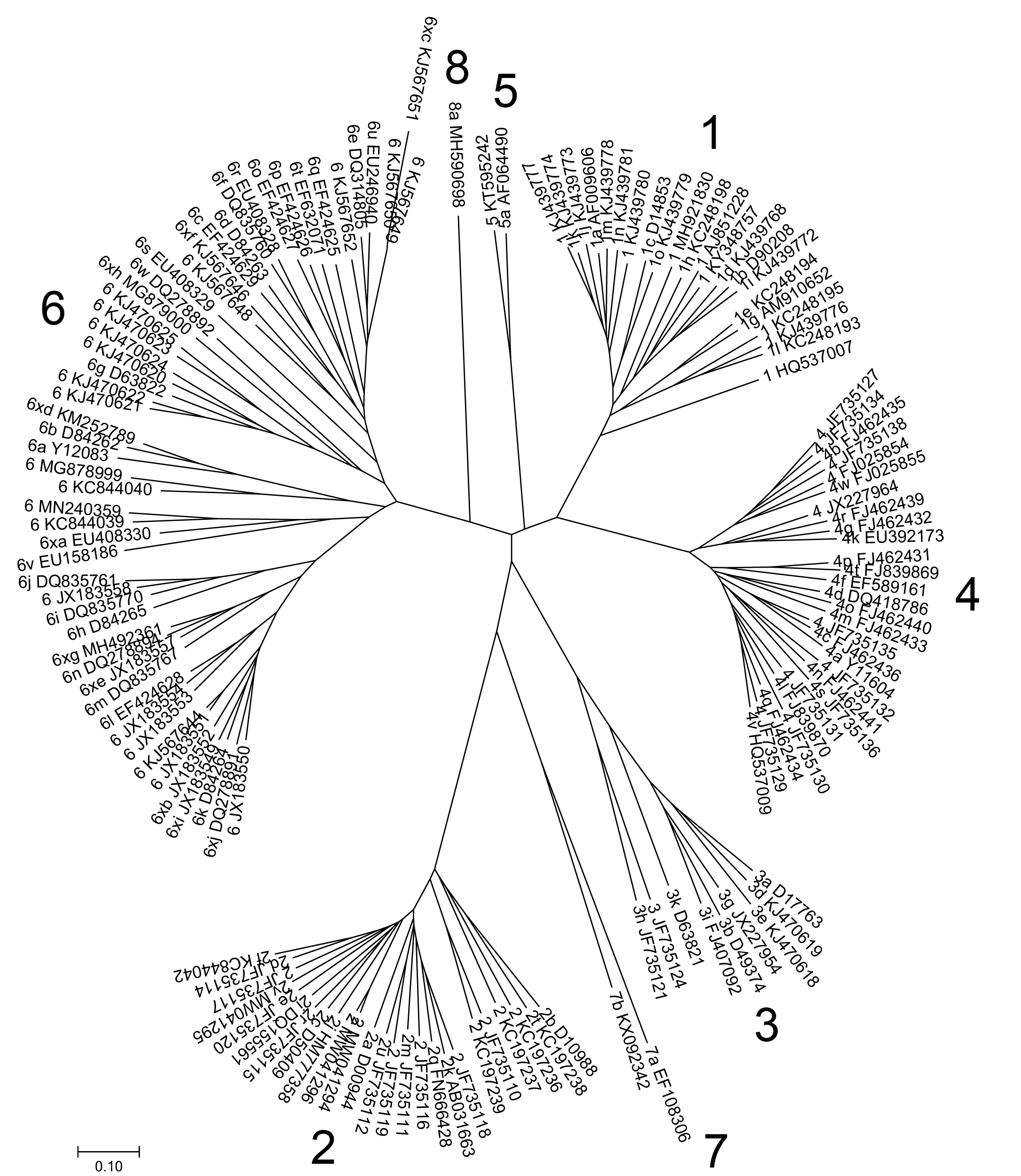
The number of confirmed genotypes and subtypes has increased from the 18 listed in 2005 (1) to 67 in 2013 (2) 86 in 2017, 90 in May 2019 and 93 in March 2022 (Table 1). A neighbour-joining phylogenetic tree of maximum composite likelihood nucleotide distances between the coding regions of these isolates, together with other isolates that have yet to be assigned a subtype is shown below. Clades corresponding to each genotype were supported by 100% of bootstrap replicates.
Rules for genotype assignments:
As proposed in the 2005 consensus proposal (1), for an HCV isolate to be considered as a new confirmed genotype or subtype, a complete coding region sequence should be obtained that:
a) Forms a distinct phylogenetic group from previously described sequences
b) Is represented by at least three epidemiologically unlinked isolates
c) Does not represent a recombinant between other genotypes or subtypes.
Novel complete coding region sequences that are represented by an insufficient number of isolates are listed in Table 2. Novel genotypes or subtypes based on partial genome sequences and/or an insufficient number of isolates will no longer be assigned provisional names; previous provisional assignments awaiting confirmation are listed in Table 3. A list of recombinant forms of HCV is listed in Table 4. Reference alignments are available as FASTA and SSE downloads.
We urge all researchers describing new variants to contact Donald Smith ([email protected]) or Peter Simmonds ([email protected]) before publishing assignments so that naming conflicts can be avoided and appropriate classifications can be made. All sequence data provided will be treated as strictly confidential.
HCV Databases. An alignment of representatives of these genotypes / subtypes (updated as new variants are assigned) is available on this site in FASTA and SSE formats or at http://hcv.lanl.gov/content/sequence/NEWALIGN/align.html. The following databases have or are in the process of updating their sequence databases, and typing tools to match the updated assignment of HCV genotypes and subtypes described here:
ICTV Flaviviridae Online Report (https://ictv.global/report/flaviviridae)
Comet (http://comet.retrovirology.lu/)
Hepatitis Virus Database (http://s2as02.genes.nig.ac.jp/)
HCV-Glue (http://hcv.glue.cvr.ac.uk/#/home)
Los Alamos Phyloplace (http://hcv.lanl.gov/content/sequence/phyloplace/)
Oxford HCV subtyping tool (http://www.viprbrc.org/brc/home.do?decorator=flav_hcv)
Virus Pathogen Resource (ViPR) (http://www.viprbrc.org/brc/home.do?decorator=flav_hcv)
References
1. Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973 (pdf available here)
2. Smith, D. B., Bukh, J., Kuiken, C., Muerhoff, A.S., Rice, C.M., Stapleton, J.T., and Simmonds, P. 2013. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes, updated criteria assignment web resource Hepatology (pdf available here)
3. Simmonds, P., Becher, P., Bukh, J., Gould, E.A., Meyers, G., Monath, T., Muerhoff, S., Pletnev, A., Rico-Hesse, R., Smith, D.B. and Stapleton, J.T., 2017. ICTV Virus Taxonomy Profile: Flaviviridae. Journal of General Virology, 98(1), pp.2-3.
4. Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics. 21:379-384
5. Combet, C., F. Penin, C. Geourjon, and G. Deleage. 2004. HCVDB : Hepatitis C Virus Sequences Database. Appl. Bioinformatics. 3:237-240
