Subfamily: Alpharhabdovirinae
Genus: Sigmavirus
Distinguishing features
Viruses assigned to the genus Sigmavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Sigmaviruses each infect dipteran insects of a single species in which they are transmitted only vertically. Sigmavirus genomes may feature an additional gene (X) located between the P and M genes, encoding a protein of unknown function.
Virion
Morphology
Bullet-shaped particles of approximately 80 nm × 100 nm with prominent surface projections have been reported for Drosophila melanogaster sigmavirus (DMelSV; species Sigmavirus melanogaster) (L'Heritier 1958, Teninges 1968, Richard-Molard et al., 1984).
Nucleic acid
Sigmavirus genomes consist of a single molecule of negative-sense, single-stranded RNA and range from approximately 11.1–14.5 kb (Teninges et al., 1993, Longdon et al., 2010, Longdon et al., 2015, Litov et al., 2021, Sharpe et al., 2021, Kamani et al., 2022).
Proteins
The N, P, M, G and L share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses (Teninges and Bras-Herreng 1987, Teninges et al., 1993, Bras et al., 1994, Landes-Devauchelle et al., 1995). The X (U1) proteins are of unknown function. They range from 224 to 321 amino acids (~25.0 kDa to ~36.9 kDa) and generally share very low or no detectable sequence homology amongst viruses assigned to different species. The DmelSV X protein has been observed to contain 3 sequence motifs that are related to conserved motifs in reverse transcriptases but the significance of this is not known (Landes-Devauchelle et al., 1995). Other likely sigmaviruses (currently unclassified) may encode other accessory proteins of unknown function (Longdon et al., 2015).
Genome organisation and replication
Sigmavirus genomes include five genes (N, P, M, G and L) encoding the structural proteins and may contain an additional long ORF between the P and M genes encoding a protein of unknown function (X or U1) (Figure 1 Sigmavirus) (Teninges et al., 1993). All ORFs occur in discrete transcriptional units including conserved transcription initiation and transcription termination/polyadenylation sequences. Variations in genome length are due primarily to significant variations in the length of 3′- and 5′-untranslated regions which may be very long (e.g., 664 nt 3′-untranslated region in the G mRNA of Drosophila affinis sigmavirus (DAffSV; species Sigmavirus affinis) (Longdon et al., 2015).
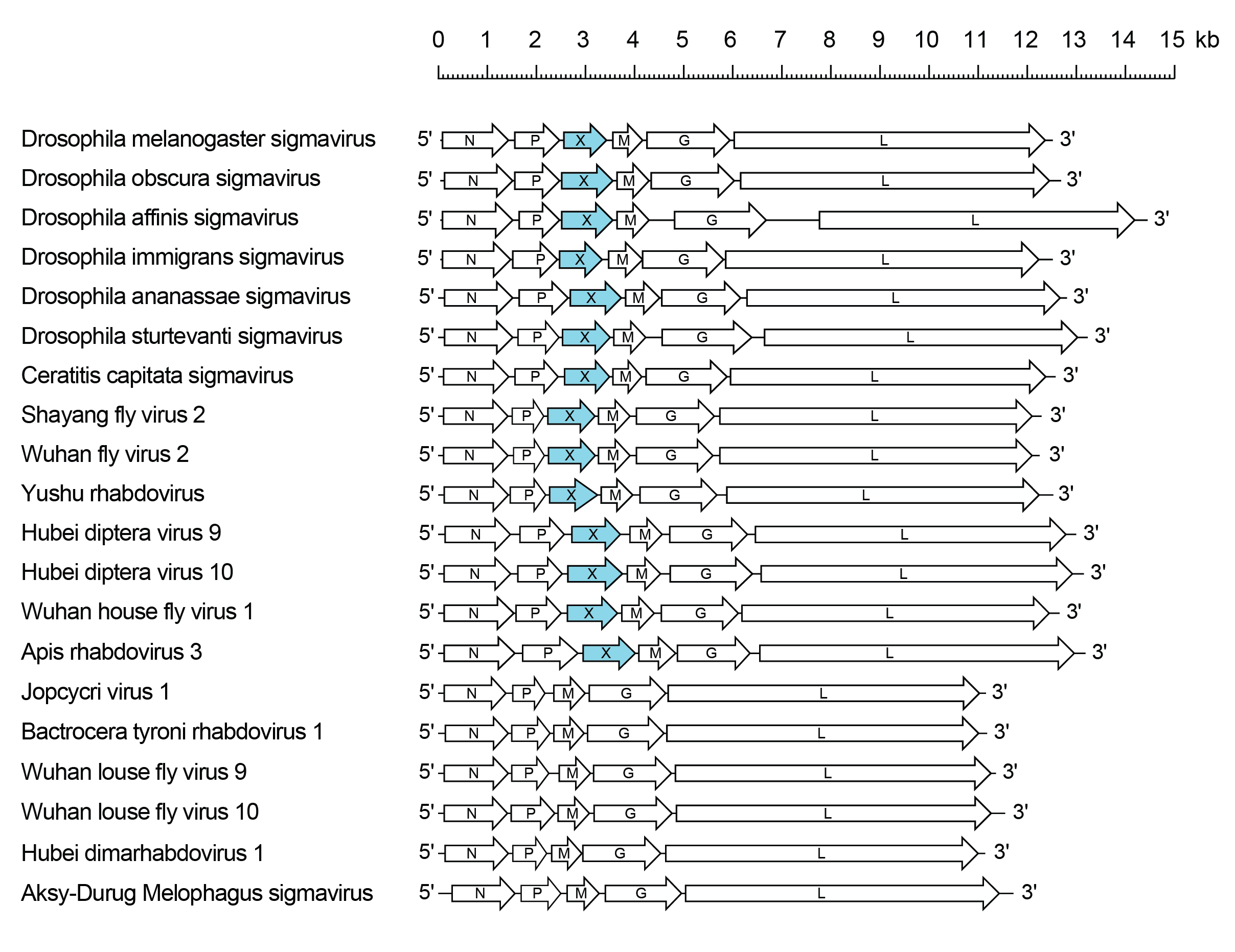 |
| Figure 1 Sigmavirus. Schematic representation of sigmavirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. ORF X (U1) encoding a protein of unknown function is highlighted (blue). |
Biology
Sigmaviruses infect insects in the order Diptera including the families Drosophilidae, Muscidae, Tephritidae and Hippoboscidae, and possibly a wider range of dipteran insects (Li et al., 2015, Longdon et al., 2015, Shi et al., 2016, Litov et al., 2021, Sharpe et al., 2021, Kamani et al., 2022). However, Apis rhabdovirus 3 (ApRV3; species Sigmavirus sichuan) was detected in eastern honey bees (Apis cerana) and Yushu rhabdovirus (YsRV; species Sigmavirus yushu) was detected in a pool of bird fecal samples, each by metagenomic sequencing. Sigmaviruses are transmitted only vertically through eggs or sperm (Fleuriet 1988, Longdon et al., 2011). Sigmaviruses have not been associated with disease but flies infected with sigmaviruses become paralysed or die upon exposure to high concentrations of carbon dioxide (L'Heritier 1958, Longdon et al., 2010); however, this property has also been associated with other rhabdoviruses (Bussereau and Contamine 1980, Rosen 1980).
Antigenicity
DmelSV has been shown to cross-react in indirect immunofluorescence tests with several rhabdoviruses including Parry Creek virus (genus Hapavirus), Humpty Doo virus (related, unclassified rhabdovirus), Tibrogargan virus (genus Tibrovirus) and Tupaia rhabdovirus (genus Tupavirus) (Calisher et al., 1989).
Species demarcation criteria
Viruses assigned to different species within the genus Sigmavirus have one or both of the following characteristics: A) minimum amino acid sequence divergence of 20% in the L protein; and B) occupy different ecological niches as evidenced by differences in hosts.
As sigmaviruses are transmitted only vertically, resulting in extreme host fidelity over ecological time scales, viruses found within different host species are likely to represent different virus species. Although host-switching does occur over evolutionary time scales, it is possible that highly related viruses could originate in hosts of different species but host-switching events are likely to be rare. Species demarcation based upon the host species should be supported by phylogenetic analysis based upon L gene sequences to indicate that the proposed virus species represents a distinct lineage and, if possible, estimates of genetic diversity to demonstrate much lower diversity within than between viruses assigned to different species. Typically, these will be <5% L amino acid sequence diversity for viruses within species and >20% diversity between species.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Bactrocera dorsalis sigmavirus | MN745080 | BDorSV |
| Drosophila algonquin sigmavirus | KR822824* | DAlgSV |
| Drosophila montana sigmavirus | KR822815* | DMonSV |
| Hangzhou rhabdovirus 4 | MZ209737 | HzRV4 |
| Scaptodrosophila deflexa sigmavirus | KR822820*; KR822821*; KR822822* | SDefSV |
| Wuhan louse fly virus 8 | KM817655* | WhLFV-8 |
Virus names and virus abbreviations are not official ICTV designations.
* Coding region sequence incomplete

