Subfamily: Alpharhabdovirinae
Genus: Mousrhavirus
Distinguishing features
Viruses assigned to the genus Mousarhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Moussa virus (MOUV; species Mousrhavirus moussa), the only virus assigned to the genus, was isolated from hard ticks.
Virion
Morphology
Bullet-shaped MOUV particles have been observed by negative-contrast electron microscopy.
Nucleic acid
The MOUV genome consist of a single molecule of negative-sense, single-stranded RNA of approximately 11.5 kb (Quan et al., 2010).
Proteins
The MOUV N, P, M, G and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses.
Genome organisation and replication
The MOUV genome includes only the five genes (N, P, M, G and L) encoding the structural protein (Figure 1 Mousrhavirus).
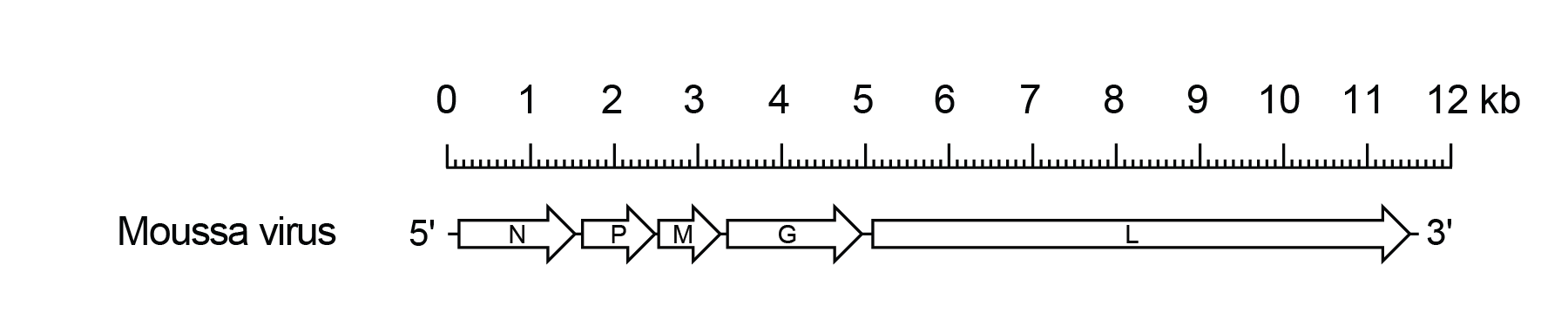 |
| Figure 1 Mousrhavirus. Schematic representation of the Moussa virus genome shown in reverse (positive-sense) polarity. The genome contains only five long open reading frames (ORFs) in the N, P, M, G and L genes (open arrows). |
Biology
MOUV was isolated on several occasions in 2004 from mosquitoes (Culex decens Theobald, 1901) collected near the Taï National Park in Côte d’Ivoire (Quan et al., 2010).
Species demarcation criteria
There is currently only a single species in the genus. For viruses to be assigned to different species within the genus, several of the following characteristics would have to be fulfilled: A) minimum amino acid sequence divergence of 10% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) they can be distinguished in virus neutralisation tests; and F) they occupy different ecological niches as evidenced by differences in vertebrate hosts and or arthropod vectors.

