Subfamily: Alpharhabdovirinae
Genus: Alphapaprhavirus
Distinguishing features
Viruses assigned to the genus Alphapaprhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Members of the genus have been detected in moths and butterflies (order Lepidoptera). They feature two consecutive genes encoding structurally related class I transmembrane glycoproteins. They are distinct phylogenetically from viruses assigned to the genus Betapaprhavirus.
Virion
Morphology
Viruses assigned to the genus have not yet been isolated or visualized by electron microscopy.
Nucleic acid
Alphapaprhavirus genomes consist of a single molecule of negative-sense, single-stranded RNA of approximately 13.1 kb (Longdon et al., 2015, Shi et al., 2016, Käfer et al., 2019).
Proteins
Alphapaprhavirus N, P, M and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses. Alphapaprhavirus encode two class I transmembrane glycoproteins (G1 and G2). Alignment with the G protein of vesicular stomatitis Indiana virus indicates that both G1 and G2 share 12 conserved cysteine residues that are typical of animal rhabdovirus G proteins (Walker and Kongsuwan 1999, Roche et al., 2006). They also share two additional cysteine residues that are predicted to form a seventh disulphide bridge.
Genome organisation and replication
Alphapaprhavirus genomes include the four genes (N, P, M and L) encoding the structural proteins (Figure 1 Alphapaprhavirus). They also include two genes (G1 and G2) encoding structurally related class I transmembrane glycoproteins that are orthologous and may have arisen by gene duplication. It is not known whether one or both glycoproteins are present in virions.
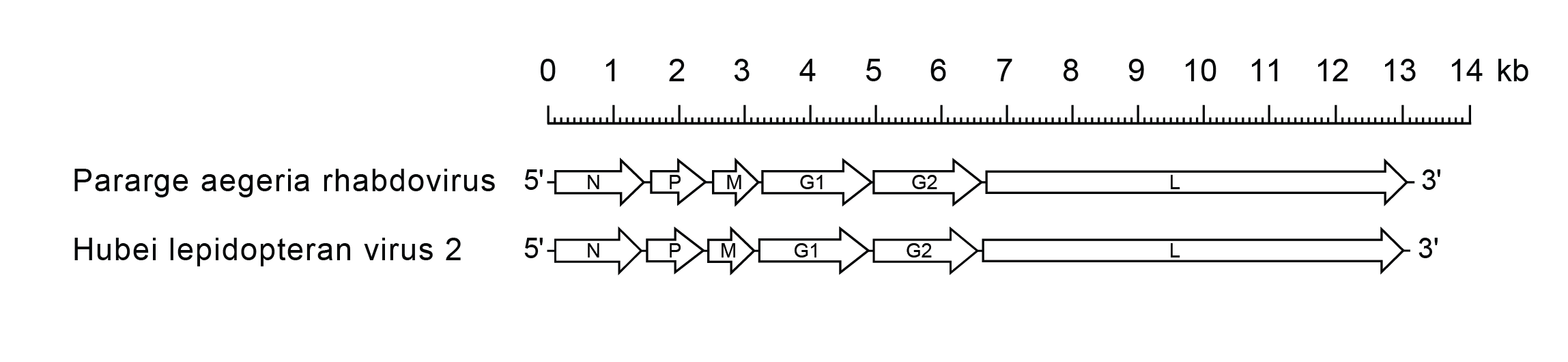 |
| Figure 1 Alphapaprhavirus. Schematic representation of genomes of alphapaprhaviruses shown in reverse (positive-sense) polarity. The six long open reading frames (ORFs) in the N, P, M, G1, G2 and L genes are shown (open arrows). |
Biology
Pararge aegeria rhabdovirus (species Alphapaprhavirus pararge) was detected in a laboratory population of butterflies (Pararge aegeria) in Belgium, in 2013 (Longdon et al., 2015). The virus was also detected as a sequence read in the transcriptome of butterflies (P. aegeria) collected in North Rhine-Westphalia, Germany, in 2011, and named lepidopteran rhabdo-related virus OKIAV12 (Käfer et al., 2019). Hubei lepidoptera virus 2 (species Alphapaprhavirus hubei) was detected in insects (order Lepidoptera) collected in Hubei Province, China, in 2013 (Shi et al., 2016). No isolates are currently available for either of these viruses.
Species demarcation criteria
Viruses assigned to different species within the genus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) can be distinguished in virus neutralisation tests; and F) occupy different ecological niches as evidenced by differences in invertebrate hosts.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Gata virus | KX852386 | GATV |
| Orgi virus | KX852388 | ORGIV |
Virus names and virus abbreviations are not official ICTV designations.

