Subfamily: Alpharhabdovirinae
Genus: Cetarhavirus
Distinguishing features
Viruses assigned to the genus Cetarhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. They have been isolated from marine cetaceans (dolphins and porpoises). They are most closely related to perhabdoviruses, sineperhaviruses, scophrhaviruses, all of which infect finfish.
Virion
Morphology
Cetarhavirus virions have bullet-shaped morphology and measure 110–145 nm in length and 70–85 nm in diameter (Osterhaus et al., 1993, Emelianchik et al., 2019).
Nucleic acid
The genome consists of a single molecule of negative-sense, single-stranded RNA of approximately 11 kb (Osterhaus et al., 1993, Emelianchik et al., 2019).
Proteins
N, P, M, G and L share significant sequence homology with the homologous proteins of other rhabdoviruses.
Genome organisation and replication
Cetarhavirus genomes include five genes in the order 3′-N-P-M-G-L-5′ encoding a nucleoprotein, polymerase-associated protein, matrix protein, glycoprotein and RNA-directed RNA polymerase, respectively (Figure 1.Cetarhavirus). The dolphin rhabdovirus (DRV; species Cetarhavirus laganorhynchus) genome contains a leader region of 66 nt preceding the transcription initiation of the N gene, and a trailer of 6 nt following the transcription termination of the L gene (Osterhaus et al., 1993). The transcriptional initiation and the termination/polyadenylation signals are conserved for all genes, 3′-UUGUC and 3′-RUAC(U)7, respectively, but the non-transcribed intergenic regions are variable. There is inverse complementarity between the 3′-leader and 5′-trailer sequences.
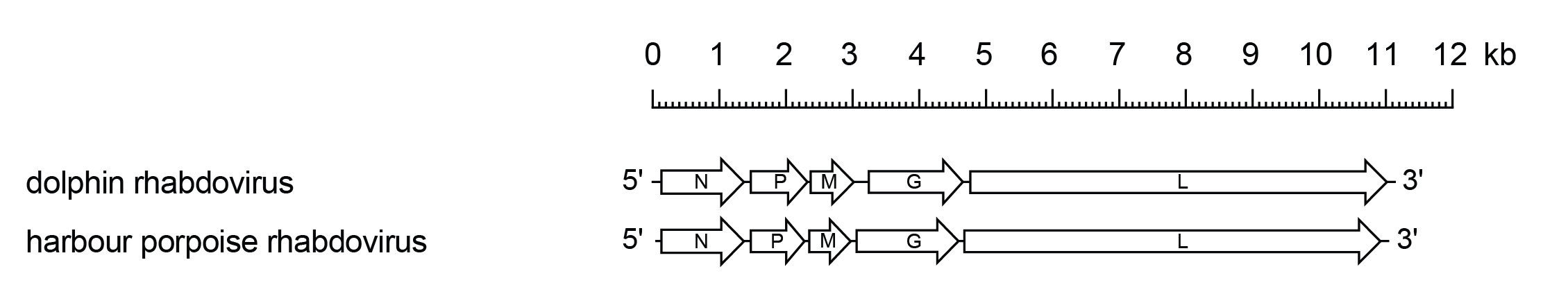 |
| igure 1.Cetarhavirus. Schematic representation of cetarhavirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. |
Biology
DRV was isolated from a white-beaked dolphin (Lagenorhynchus albirostris) stranded on the Dutch island of Schiermonnikoog in 1992 (Osterhaus et al., 1993). It was shown to replicate in African green monkey kidney (Vero) cells. Neutralising antibody was detected in various cetaceans (dolphins, porpoises, whales) and pinnipeds (seals) sampled from the coast of northwest Europe or the Mediterranean Sea (Osterhaus et al., 1993, Siegers et al., 2014). Harbour porpoise rhabdovirus (species Cetarhavirus phocoena) was isolated from a harbour porpoise (Phocoena phocoena) stranded off the coast of Alaska in 2013 (Emelianchik et al., 2019). It was shown to replicate in primary beluga whale (Delphinapterus leucas) kidney cells.
Antigenicity
DRV failed to cross-react with rabies virus, bovine ephemeral fever virus or several vesiculoviruses in indirect immunofluorescence or virus neutralisation tests (Osterhaus et al., 1993).
Species demarcation criteria
Viruses assigned to different species within the genus Cetarhavirus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in N proteins; B) minimum sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in G proteins; D) can be distinguished in virus neutralisation tests; and E) occupy different ecological niches as evidenced by differences in vertebrate hosts.

