Subfamily: Betarhabdovirinae
Genus: Varicosavirus
Distinguishing features
Viruses assigned to the genus Varicosavirus form a monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Varicosaviruses infect plants and have bi-segmented genomes. Lettuce big vein-associated virus (LBVaV; species Varicosavirus lactucae) has non-enveloped, flexuous, rod-shaped virions that, in electron micrographs, resemble nucleocapsids of other rhabdoviruses.
Virion
Morphology
LBVaV vrions are fragile, non-enveloped rods mostly measuring 324–152 nm × approximately18 nm; each has a central canal approximately 3 nm in diameter and an obvious helix with a pitch of about 5 nm (Figure 1 Varicosavirus). The helix of particles, especially those in purified preparations, tends to loosen and particles are then seen as partially uncoiled filaments (Kuwata and Kubo 1981, Kuwata et al., 1983, Vetten et al., 1987).
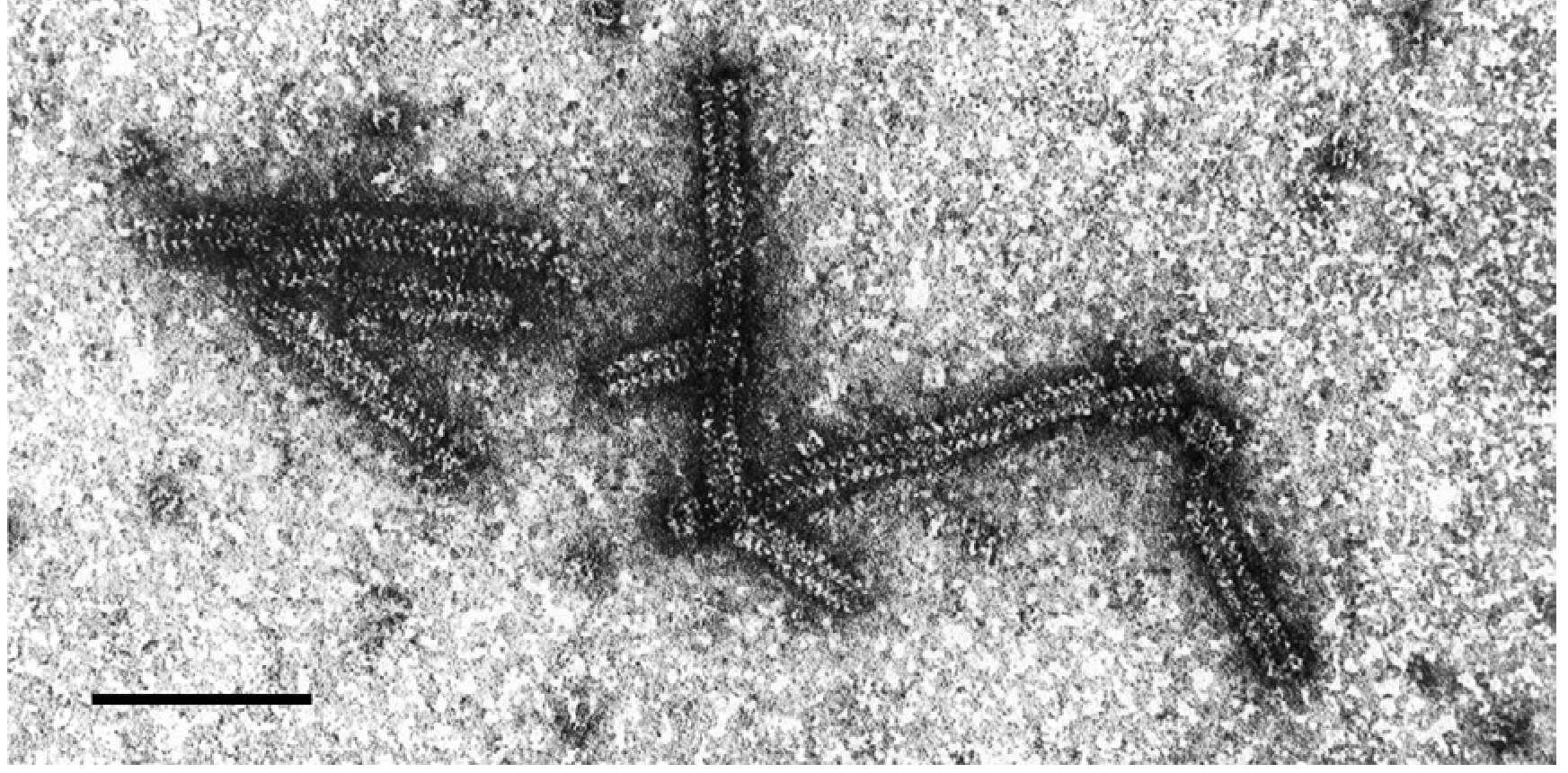 |
| Figure 1 Varicosavirus. Negative-contrast electron micrograph of virions of an isolate of lettuce big vein-associated virus. The bar represents 100 nm (Courtesy J.A. Walsh and C.M. Clay.) |
Physicochemical and physical properties
Virus particles have a density in Cs2SO4 of about 1.27 g cm-3 (Kuwata et al., 1983).
Nucleic acid
LBVaV, Alopecurus myosuroides varicosavirus 1 (AMVV1; species Varicosavirus alopecuri), Allium angulosum virus 1 (AAnV1; species Varicosavirus allii), Brassica rapa virus 1 (BrRV1; species Varicosavirus brassicae), morning glory varicosavirus (MGVV; species Varicosavirus ipomoeae), Lolium perenne virus 1 (LoPV1; species Varicosavirus lolii), Melampyrum roseum virus 1 (MelRoV1; species Varicosavirus melampyri), , Vitis varicosavirus (VVV; species Varicosavirus vitis), Xinjiang varicosavirus (XVV; species Varicosavirus xinjiangense), Zostera-associated varicosavirus 1 (ZaVV1; species Varicosavirus zosterae), red clover associated varicosavirus (RCaVV; species Varicosavirus trifolii), and the thirty-two varicosavirus recently identified (Bejerman et al., 2022) have negative-sense, single-stranded RNA genomes encapsidated by N (previously termed coat protein, CP). The genomes of varicosaviruses (10.4–12.9 kb) occurs as two segments of 6.8 kb (RNA1) and 4.0–6.1 kb (RNA2) (Sasaya et al., 2002, Sasaya et al., 2004, Bejerman et al., 2021, Bejerman et al., 2022). The 3′- and 5′-terminal sequences of the two genome segments are similar but do not exhibit inverse complementarities. Compared with orchid fleck virus (OFV; species Dichorhavirus orchidaceae), purified LBVaV virions contain relatively large amounts of anti-genomic RNAs (Sasaya et al., 2001). Conserved motifs in the transcription initiation and termination signals resemble those of viruses in the genus Cytorhabdovirus.
Proteins
LBVaV RNA2 encodes the 397-aa N with a predicted mass of 44.5 kDa (Mr approximately 48 kDa), as well as protein 2 (36 kDa), protein 3 (32 kDa), protein 4 (19 kDa) and protein 5 (41 kDa) which are of unknown functions but thought to be equivalent to P, P3, M and G of cytorhabdoviruses (Sasaya et al., 2004, Kormelink et al., 2011). LBVaV RNA1 encodes protein 6 (5 kDa), also of unknown function, and the 232 kDa L (Sasaya et al., 2002). N and L share a moderate amount of amino acid sequence identity with their counterparts in other rhabdoviruses.
Genome organisation and replication
All varicosaviruses have bi-segmented genomes and contain only the L gene in RNA1, except for LBVaV in which RNA1 contains two genes in the order 3′-6-L-5. LBVaV, VVV, XVV and other seven viruses. RNA2 contains five genes in the order 3′-N-2-3-4-5-5′, one or two genes more than the other viruses of the genus (Figure 2 Varicosavirus). Although the genome organisation of LBVaV is similar to that of other rhabdoviruses, LBVaV does not have a 3′-non-coding leader sequence. Capped and polyadenylated monocistronic mRNAs are transcribed from individual genes.
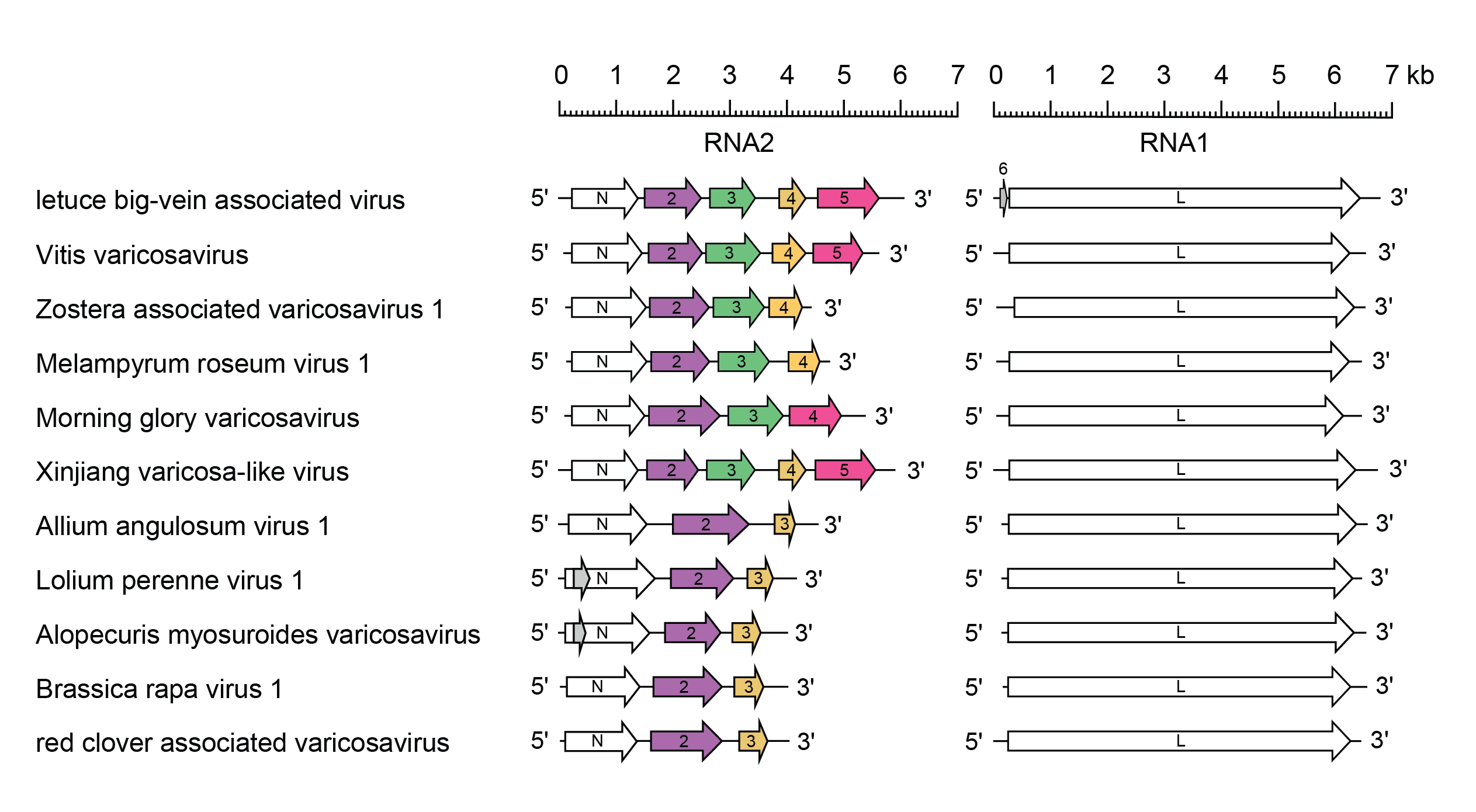 |
| Figure 2 Varicosavirus. Schematic representation of a selection of varicosavirus genomes (RNA1 and RNA2) shown in reverse (positive-sense) polarity. N and L represent ORFs encoding the known structural proteins. Other ORFs (2, 3, 4 and 5) encode putative proteins of unknown function. ORFs that appear to be homologous are shown in the same colour. Alternative ORFs in the N genes (grey) may or may not be expressed. In addition to those genomes shown: A) Arceuthobium virus 8, Artemisia virus 1, Centaurea virus 1, Erysimum virus 1, Leucanthemum virus 1, Melilotus virus 1, Pennisetum virus 1, Raphanus virus 1, Silene virus 1, Tanacetum virus 1 and Triticum virus 1 have 3 genes in the RNA 2 like Allium angulosum virus 1, Brassica rapa virus 1 and red clover-associated varisocavirus; B) Aponogeton virus 1, Brassica virus 2, Caladenia virus 1, Cucumis virus 1, Frullania virus 1, Guizotia virus 1, Holcus virus 1, Luffa virus 1, Monoclea gottschei varicosa-like virus, Ribes virus 1, Streptoglossa virus 1, Treubia virus 1, Vincetoxicum virus and Zea virus 1 have 4 genes in the RNA 2 like Melampyrum roseum virus 1, morning glory varicosavirus and Zostera associated virus 1; and C) Aconitum virus 1, Apera virus 1, Asclepias syriaca virus 3, Didymochlaena virus 1, Primula virus 1, Ranunculus virus 1, tree-fern varicosa-like virus have 5 genes in the RNA 2 like vitis varicosavirus and Xinjiang varicosa-like virus. |
Biology
LBVaV and tobacco stunt virus (TStV; currently unclassified) are transmitted in soil and in hydroponic systems by zoospores of the chytrid fungus Olpidium virulentus (a noncrucifer strain of Olpidium brassicae) (Campbell 1962, Hiruki 1967, Sasaya and Koganezawa 2006, Maccarone et al., 2010b). LBVaV is frequently associated with lettuce big vein disease that is caused by Mirafiori lettuce big-vein virus (MLBVV; species Ophiovirus mirafioriense, family Aspiviridae) (Roggero et al., 2000, Lot et al., 2002). Due to the longevity of infectious LBVaV from stored O. virulentus resting spores, it is assumed that the virus is carried internally by the fungus. The viruses are also transmitted experimentally, sometimes with difficulty, by mechanical inoculation (Huijberts et al., 1990). Neither of the viruses is reported to be seed-transmitted. LBVaV has been reported from Europe, United States, Japan, Saudi Arabia and Australia (Navarro et al., 2005, Maccarone et al., 2010a, Alemzadeh and Izadpanah 2012, Umar et al., 2017).
Antigenicity
N does not induce a strong antibody response in laboratory animals. LBVaV is closely related serologically to TStV, which is currently unclassified but may be considered to be a strain of LBVaV (Kuwata and Kubo 1986, Sasaya et al., 2005).
Species demarcation criteria
Viruses assigned to different species within the genus Varicosavirus have several of the following characteristics: A) the nucleotide sequence identity of the L ORF is lower than 75%; B) they occupy different ecological niches as evidenced by differences in hosts and/or chytrid vectors; and C) they can be clearly distinguished in serological tests or by nucleic acid hybridisation.
Related, unclassified viruses
| Virus name | Accession number
| Virus abbreviation
|
| tobacco stunt virus | AB190521* | TStV |
Virus names and virus abbreviations are not official ICTV designations.
* Coding region sequence incomplete

