Subfamily: Alpharhabdovirinae
Genus: Ohlsrhavirus
Distinguishing features
Viruses assigned to the genus Ohlsrhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Viruses assigned to the genus have been discovered in culicine mosquitoes (primarily Ochlerotatus spp. or Culex spp.) collected from Europe, Asia, Australia, Africa or the Americas.
Virion
Morphology
Virion morphology has not been described.
Nucleic acid
Ohlsrhavirus genomes consist of a single molecule of negative-sense, single-stranded RNA of approximately 11.5–13.4 kb (Coffey et al., 2014, Hang et al., 2016, Reuter et al., 2016, Shahhosseini et al., 2017, Shi et al., 2017).
Proteins
Ohlsrhavirus N, P, M, G and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses.
Genome organisation and replication
Ohlsrhavirus genomes include only five genes (N, P, M, G and L) encoding the structural proteins (Figure 1 Ohlsrhavirus).
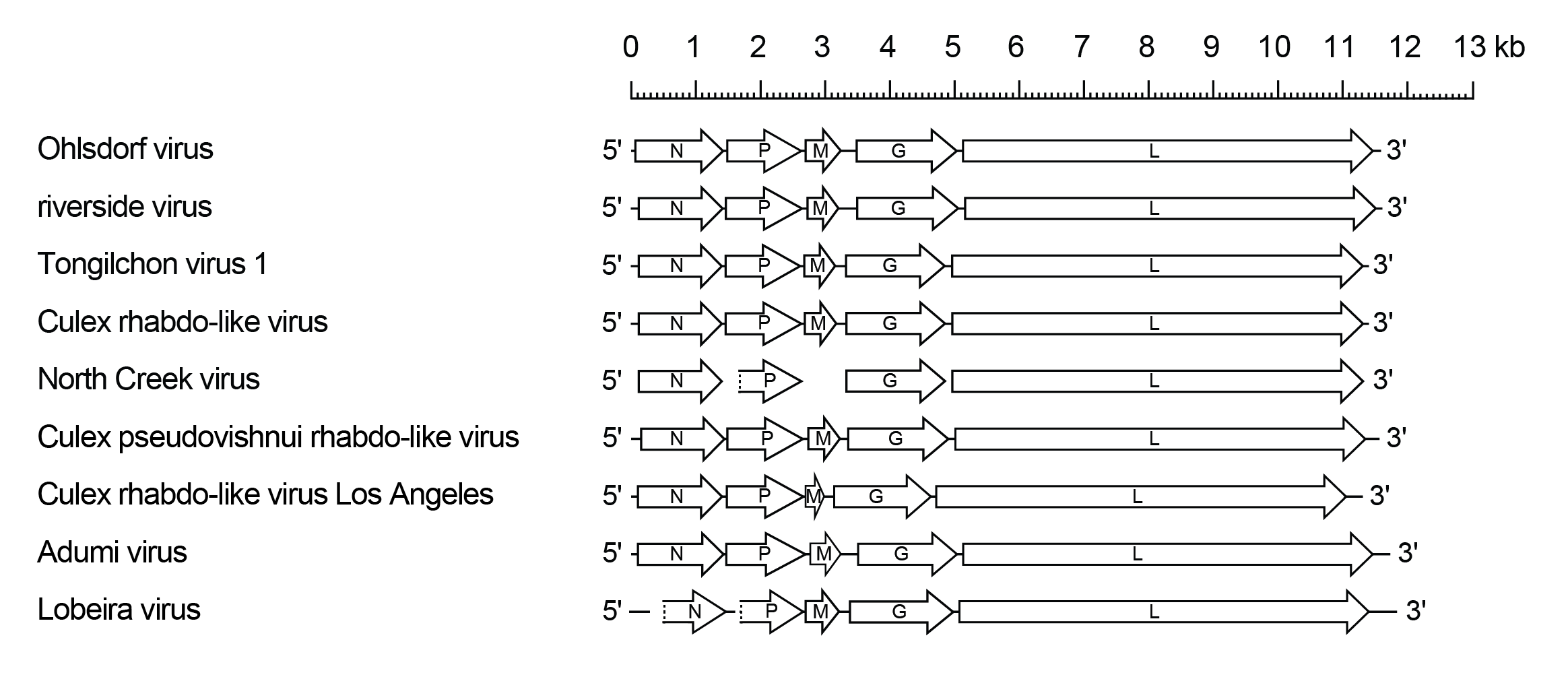 |
| Figure 1 Ohlsrhavirus. Schematic representation of ohlsrhavirus genomes shown in reverse (positive-sense) polarity. Each genome contains long open reading frames (ORFs) in the N, P, M, G and L genes (open arrows). In North Creek virus, sequences are not yet available for the M gene or the start of the P gene. |
Biology
Ohlsrhaviruses have been detected by high-throughput sequencing in culicine mosquitoes (Culicidae) collected in Europe, Asia, Australia or the Americas. Ohlsdorf virus (OHSDV; species Ohlsrhavirus ohlsdorf) was first detected in mosquitoes (Ochlerotatus cantans) collected from Hamburg, Germany, in 2012, and subsequently in mosquitoes of the same species collected from other parts of Germany from 2013-2015 (Shahhosseini et al., 2017). Riverside virus (RISV; species Ohlsrhavirus riverside) was detected in three pools of mosquitoes (Ochlerotatus sp.) collected from near the Danube River and Drava River in Hungary, in 2014 (Reuter et al., 2016). Tongilchon virus 1 (TCHV1; species Ohlsrhavirus tongilchon) was detected in mosquitoes (Culex bitaeniorhynchus) collected in the Republic of Korea, in 2012 (Hang et al., 2016). North Creek virus (NORCV; species Ohlsrhavirus northcreek) was detected in mosquitoes (Cx. sitiens) collected in New South Wales, Australia, in 1997 (Coffey et al., 2014). Culex rhabdo-like virus (CRLV; species Ohlsrhavirus culex) was detected in mosquitoes (Cx. quinquefasciatus, Cx. globocoxitus and Cx. australicus) collected in Western Australia, in 2015 (Shi et al., 2017). Culex pseudovishnui rhabdo-like virus (CpRLV; species Ohlsrhavirus pseudovishnui) was detected in mosquitoes (Cx. pseudovishnui) collected in Japan, in 2017 (Faizah et al., 2020). Culex rhabdo-like virus Los Angeles (CRLVLA; species Ohlsrhavirus angeles) was detected in mosquitoes (Culex sp.) collected in California, in 2016 (Sadeghi et al., 2018). Lobeira virus (LOBV; species Ohlsrhavirus lobeira) was detected in mosquitoes (Psorophora albigenu) collected in Brazil, in 2014 (Lara Pinto et al., 2017, Maia et al., 2019). Adumi virus (ADUMV; species Ohlsrhavirus adumi) was detected in mosquitoes (species not recorded) collected in Uganda. No virus isolates are currently available. No information is currently available on possible vertebrate hosts.
Species demarcation criteria
Viruses assigned to different species within the genus Ohlsrhavirus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) they can be distinguished in virus neutralisation tests; and F) they occupy different ecological niches as evidenced by differences in vertebrate hosts and or arthropod vectors.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Culex quinquefasciatus rhabdo-like virus 1 | OQ067689 | CqRLV1 |
| Culex rhabdo-like virus 2 | OQ067690 | CRLV2 |
| dipteran rhabdo-related virus 5 | TSA#* | DiRRV5 |
| Rhabdoviridae sp. isolate XZS177686 | MW826481 |
Virus names and virus abbreviations are not official ICTV designations.
* Coding region sequence incomplete
# TSA – transcriptome shotgun assembly. See doi.org/10.5061/dryad.87vt6hm and (Käfer et al., 2019)

