Subfamily: Alpharhabdovirinae
Genus: Merhavirus
Distinguishing features
Viruses assigned to the genus Merhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences. Members of the genus have been detected culicine mosquitoes (Culicidae). Uniquely amongst rhabdoviruses reported to date, one member of the genus (Culex tritaeniorhynchus rhabdovirus; CTRV; species Merhavirus tritaeniorhynchus) has been shown to replicate in the nucleus and employ the host RNA splicing machinery.
Virion
Morphology
Enveloped rod-shaped virions of CTRV have been observed in culture supernates and thin sections of mosquito (Aedes albopictus) cell cultures (Kuwata et al., 2011). Virions were reported to be approximately 60 nm in diameter and ranged in length from 250–1,100 nm. Surface projects (6–8 nm) and an internal helical structure with a diameter of 40 nm and a pitch of 7.5– 8.0 nm (Kuwata et al., 2011).
Nucleic acid
Merhavirus genomes consist of a single molecule of negative-sense, single-stranded RNA of approximately 11.3–11.8 kb (Kuwata et al., 2011, Charles et al., 2016, Ergünay et al., 2017, Sadeghi et al., 2018).
Proteins
Merhavirus N, P, M, G and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses. Merhavirus G proteins are class I transmembrane glycoproteins. Alignment with the G protein of vesicular stomatitis Indiana virus indicates likely conservation all 12 conserved cysteine residues that are typical of animal rhabdovirus G proteins.
Genome organisation and replication
Merhavirus genomes include the five genes (N, P, M, G and L) encoding the structural proteins (Figure 1 Merhavirus). Uniquely, in CTRV, there are two ORFs in the L gene (L1 and L2) separated by an intron of 76 nt which contains the typical motif for eukaryotic spliceosomal intron-splice donor/acceptor sites (GU-AG), a predicted branch point and a polypyrimidine tract. CTRV replicates in the nucleus and utilizes the host nuclear splicing machinery (Kuwata et al., 2011).
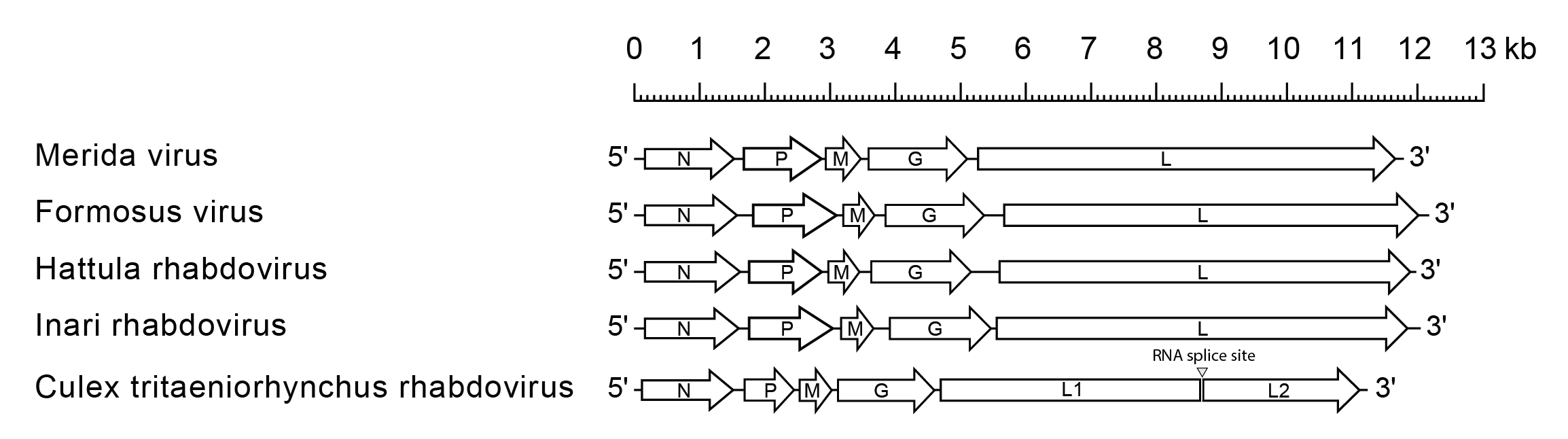 |
| Figure 1 Merhavirus. Schematic representation of merhavirus genomes shown in reverse (positive-sense) polarity. The five long open reading frames (ORFs) in the N, P, M, G and L genes are shown (open arrows). In CTRV, the L gene contains two long ORFs (L1 and L2) separated by a 76 nt intron. |
Biology
Merida virus (MERDV; species Merhavirus merida) was detected in mosquitoes (Culex quinquefasciatus) collected in Yucatan Peninsula, Mexico, in 2007 (Charles et al., 2016). The virus has also been detected in culicine mosquitoes of other species (Ochlerotatus taeniorhynchus and O. trivittatus) (Charles et al., 2016). MERDV was also detected in mosquitoes (Culex pipiens) collected in Thrace, Turkey, in 2015, and named Merida-like virus Turkey (Ergünay et al., 2017, Öncü et al., 2018). The virus was also detected in a mosquito (Culex sp.) collected in California, USA, in 2016 and named culex rhabdovirus (Sadeghi et al., 2018). Due to high levels of amino acid sequence identity, these are all considered to be variants of the same virus. No isolate of the virus is currently available.
CTRV was isolated from mosquitoes (Culex tritaeniorhynchus) collected in Chiba, Japan, and passaged in mosquito (Aedes albopictus) cell cultures (Kuwata et al., 2011). Formosus virus (FORMV; species Merhavirus formosus) was detected by metagenomic sequencing of mosquitoes (Aedes aegypti) from a laboratory colony originating from Bundibugyo, Uganda (Parry et al., 2021). Hattula rhabdovirus (HTTRV; species Merhavirus hattula) was detected by metagenomic sequencing of mosquitoes (Ochlerotatus communis) collected in Ilomantsi, Finland (Truong Nguyen et al., 2022). Inari rhabdovirus (INARV; species Merhavirus inari) was detected by metagenomic sequencing of mosquitoes (Ochlerotatus communis) collected in Lappi, Finland (Truong Nguyen et al., 2022).
Species demarcation criteria
Viruses assigned to different species within the genus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) can be distinguished in virus neutralisation tests; and F) occupy different ecological niches as evidenced by differences in invertebrate hosts.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Armigeres subalbatus rhabdovirus | LC775065 | ASubRV |
| Cambodia Anopheles rhabdovirus | OR479699 | CamAnRV |
| Evros rhabdovirus 2 | MW520372* | EVRV2 |
| Mopeia rhabdovirus | KX245155* | MOPRV |
Virus names and virus abbreviations are not official ICTV designations.
* Coding region sequence incomplete

