Subfamily: Betarhabdovirinae
Genus: Alphanucleorhabdovirus
Distinguishing features
Historically, the genera Nucleorhabdovirus and Cytorhabdovirus were established based on the sites of virus replication and morphogenesis, with nucleorhabdoviruses replicating and maturing in the nuclei of infected cells. Recently, a reclassification became necessary as phylogenetic analyses of new plant rhabdovirus genomes consistently showed that nucleorhabdoviruses did not form a monophyletic clade upon analysis of complete L protein sequence alignments (Kuhn et al., 2020).
Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences, viruses classified in the genus Alphanucleorhabdovirus form a monophyletic cluster clearly distinguished from beta-, delta- and gammanucleorhabdoviruses, and from dichorhaviruses, as well as from other plant rhabdoviruses.
Virion
Morphology
Enveloped virions are bacilliform, 45–100 nm in diameter and 130–300 nm long (Goodin and Jackson 2002, Jackson et al., 2005a).
Physicochemical and physical properties
Virus particles sediment at 800–1000 S in sucrose gradients and the buoyant density of virions is 1.18 g cm−3 in isopycnic sucrose gradients (Goodin and Jackson 2002).
Nucleic acid
The negative-sense, single-stranded RNA genome of 12.0–14.5 kb is unsegmented. Six or seven mRNAs, one for each of the encoded proteins identified in infected plants.
Proteins
N, P, M, G and L represent the five canonical rhabdovirus structural proteins. Rice yellow stunt virus (RYSV; species Alphanucleorhabdovirus oryzae) P6 has RNA silencing suppressor activity (Jackson et al., 2005a, Guo et al., 2013). The RYSV P3 protein, encoded by an ORF between the P and M ORFs, has been shown to have cell-to-cell movement activity in a heterologous virus trans-complementation assay (Huang et al., 2005). The Y proteins of potato yellow dwarf virus (PYDV; species Alphanucleorhabdovirus tuberosum) and eggplant mottled dwarf virus (EMDV; species Alphanucleorhabdovirus melongenae), also encoded by an ORF between the P and M ORFs, are thought to be movement proteins (Jackson et al., 2005a, Bandyopadhyay et al., 2010, Pappi et al., 2013).
PYDV M can induce the intranuclear accumulation of the inner nuclear membrane in the absence of any other viral protein (Bandyopadhyay et al., 2010). Protein interaction studies for PYDV in live plants have identified binary interactions between N:N, N:P, N:M, M:M, M:Y, M:G, G:G and Y:Y (Bandyopadhyay et al., 2010).
Lipids
The lipoprotein envelope is derived from the host plant or insect vector (Jackson et al., 2005a). Lipid composition is unknown.
Genome organisation and replication
The PYDV genome (12.9 kb) contains seven genes in the order 3′-N-X-P-Y-M-G-L-5′, which likely encode the nucleocapsid protein (N), phosphoprotein (polymerase cofactor) (P), movement protein (Y), matrix protein (M), glycoprotein (G) and RNA-directed RNA polymerase (L), respectively (Figure 1 Alphanucleorhabdovirus). The function of the protein encoded in the X gene has not been determined. The genome organisation of EMDV, Joa yellow blotch associated virus (JYBaV; species Alphanucleorhabdovirus joa), Physostegia chlorotic mottle virus (PhCMoV; species Alphanucleorhabdovirus physostegiae) and tomato alphanucleorhabdovirus 1 (TARV1; species Alphanucleorhabdovirus lycopersici) resembles that of PYDV. The genome of Xinjiang alphanucelorhabdovirus (XARV; species Alphanucleorhabdovirus xinjianensis) has a gene order similar to that of PYDV, except for the presence of an additional small gene (P5) between the M and G genes. The PYDV coding sequences are flanked by a 3′-leader RNA of 149 nt and a 5′-trailer RNA of 97 nt, with genes separated by conserved intergenic “gene junction” regions that are similar in both length and sequence to those of other rhabdoviruses.
The genomes of RYSV and wheat yellow striate virus (WYSV; species Alphanucleorhabdovirus tritici) have gene orders similar to that of maize mosaic virus (MMV; species Alphanucleorhabdovirus maydis), except for the presence of an additional small gene P6 between the G gene and the L gene, which in RYSV encodes a virion-associated protein. The genomes of maize Iranian mosaic virus (MIMV; species Alphanucleorhabdovirus zeairanense), taro vein chlorosis virus (TaVCV; species Alphanucleorhabdovirus colocasiae) and Morogoro maize-associated virus (MMaV; species Alphanucleorhabdovirus morogoromaydis), peach virus 1 (PeV1; species Alphanucleorhabdovirus pruni), agave tequilana virus 1 (ATV1; species Alphanucleorhabdovirus agavis) and Artemisia capillaris nucleorhabdovirus 1 (ArtCaNV1; species Alphanucleorhabdovirus artemisiae) are approximately 12.0–14.5 kb with a gene order resembling that of MMV.
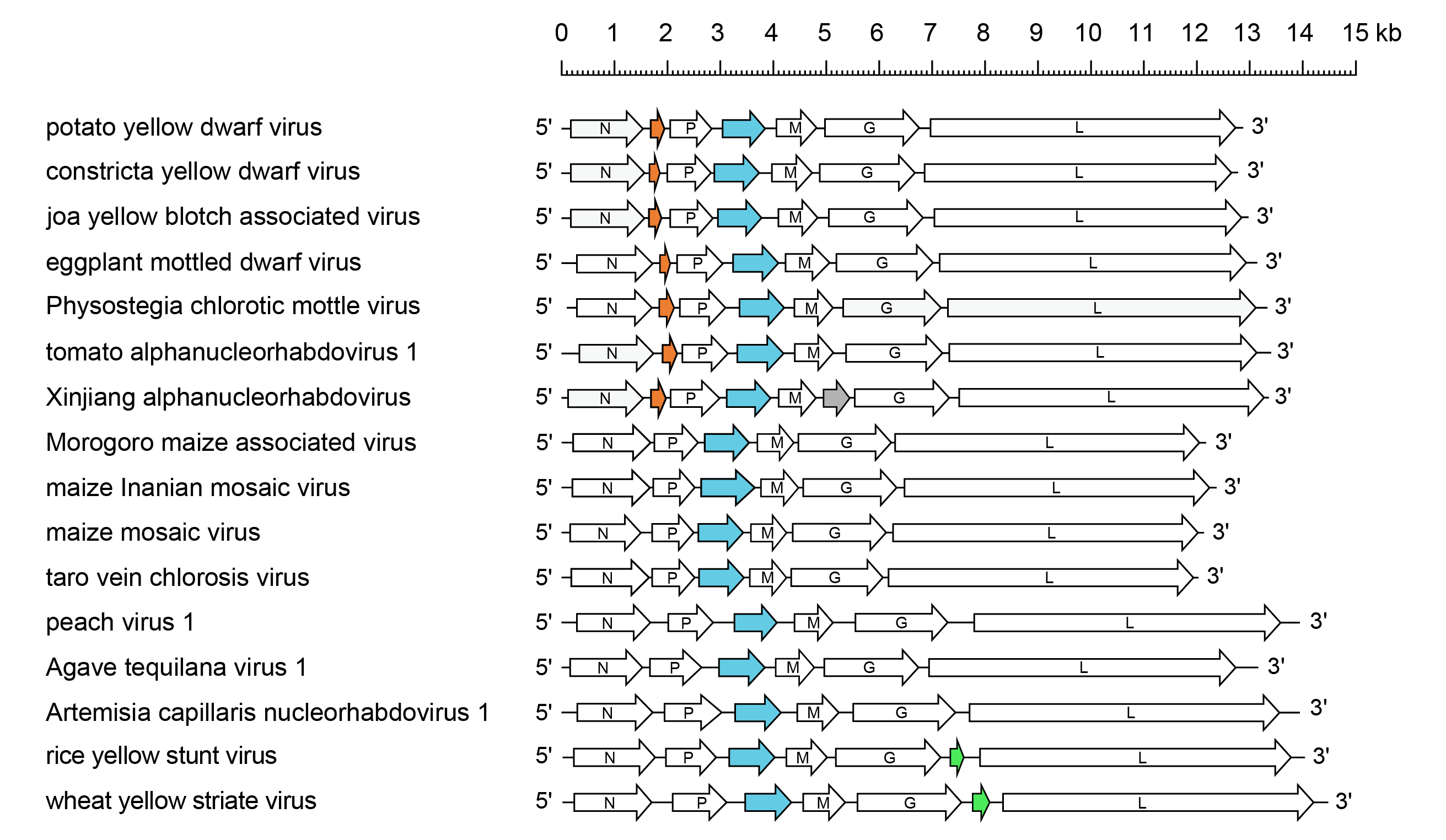 |
| Figure 1 Alphanucleorhabdovirus. Schematic representation of alphanucleorhabdovirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. ORFs encoding putative cell-to-cell movement proteins are highlighted (blue). Other ORFs encode putative accessory proteins of unknown function, including sets of small homologous proteins located between the N gene and P gene (orange), and between the G gene and L gene (green). Xinjiang alphanucleorhabdovirus encodes an additional ORF (grey). |
Alphanucleorhabdoviruses replicate in the nuclei of plant cells, which become greatly enlarged and develop large granular nuclear inclusions that are thought to be sites of virus replication (Jackson et al., 2005a).
Biology
A wide variety of monocot and dicot plants are susceptible to infection by alphanucleorhabdoviruses although each virus usually has a restricted host range (Goodin and Jackson 2002, Jackson et al., 2005a). Alphanucleorhabdoviruses are transmitted by leafhoppers (RYSV, WYSV, EMDV, PYDV) or planthoppers (MMV, MIMV). Some viruses are also transmitted during vegetative propagation, and some can be transmitted mechanically from infected sap. In all carefully examined cases, viruses replicate in cells of the insect vector as well as in the plant host (Jackson et al., 2005a).
Species demarcation criteria
Viruses assigned to different species within the genus Alphanucleorhabdovirus have several of the following characteristics: A) the nucleotide sequence identity of complete genomes is lower than 75%; B) they occupy different ecological niches as evidenced by differences in hosts and/or arthropod vectors; and C) they can be clearly distinguished in serological tests or by nucleic acid hybridisation.
Alphanucleorhabdovirus species are primarily differentiated by plant host range and vector specificity of the virus. Nucleic acid hybridisation has been used to provide confirmation of identification and serological criteria have enabled verification of common viruses that infect different hosts. However, no alphanucleorhabdovirus species have been defined unambiguously using serology. Complete genome nucleotide sequences are available for all 14 viruses currently assigned to species in the genus. RT-PCR-based assays and fluorescent viral protein localisation have proven to be useful tools for species demarcation. Hybridisation using cloned probes and RT-PCR has been used to detect the viruses.
Related, unclassified viruses
| Virus name | Accession number
| Virus abbreviation
|
| babaco nucleorhabdovirus 1 | OQ256237 | BabRV1 |
| cynodon rhabdovirus | EU650683* | CyRV |
| papaya nucleorhabdovirus 1 | MN203193* | PNRV1 |
Virus names and virus abbreviations are not official ICTV designations.
* Coding region sequence incomplete

