Subfamily: Alpharhabdovirinae
Genus: Barhavirus
Distinguishing features
Viruses assigned to the genus Barhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences. Viruses assigned to the genus have been isolated from culicine mosquitoes. There is evidence of infection in mammals, including humans. They are most closely related to rhabdoviruses in the genera Lostrhavirus and Zarhavirus, the members of which have been detected in ticks.
Virion
Morphology
Bahia Grande virus (BGV; species Barhavirus bahia) and Muir Springs virus (MSV; species Barhavirus muir) each have typical rhabdovirus-like virion morphology (60 nm × 149 nm) by transmission electron microscopy in thin sections of infected cells (Kerschner et al., 1986).
Nucleic acid
Barhavirus genomes consist of a single molecule of negative-sense, single-stranded RNA of approximately 12.6 kb (Walker et al., 2015).
Proteins
Barhavirus N, P, M, G and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses. The putative BGV Gx protein (8.0 kDa) has the structural characteristics of a class 1a viroporin featuring a short N-terminal domain, a central transmembrane domain and a highly basic C-terminal domain, and this structural arrangement is preserved in the corresponding sequence of MSV (see below). The putative BGV Nx protein (7.6 kDa) features 10 conserved cysteine residues in the central domain.
Genome organisation and replication
Barhavirus genomes include five genes (N, P, M, G and L) encoding the structural proteins (Walker et al., 2015) (Figure 1 Barhavirus). In BGV, alternative ORFs occur in the N gene (Nx) and also in the G gene overlapping the end of the G ORF (Gx). It is not known if these alternative ORFs are functional, but they are preserved in two sequenced isolates of BGV and in Harlingen virus (HARV), a strain of BGV. In MSV, there is no alternative ORF in the N gene; an alternative ORF overlapping the end of the G ORF does occur but there is no initiation codon in the published sequence.
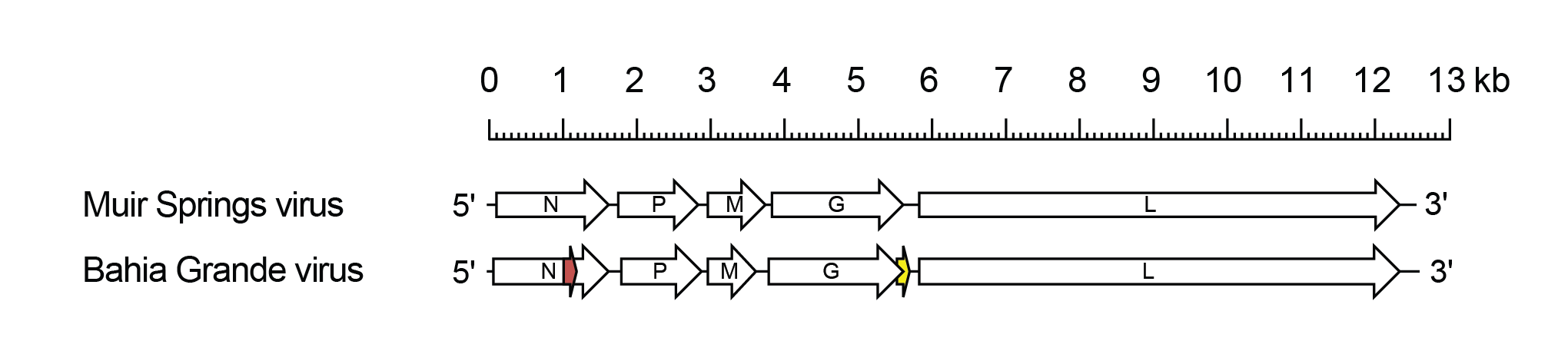 |
| Figure 1 Barhavirus. Schematic representation of barhavirus genomes shown in reverse (positive-sense) polarity. Each genome contains long open reading frames (ORFs) corresponding to the N, P, M, G and L genes (open arrows). In Bahia Grande virus, there is an alternative ORF (>180 nt) in the N gene and an an alternative ORF (Gx) encoding a viroporin-like protein occurs at the end of the G gene (yellow). |
Biology
Barhaviruses have been isolated from culicine mosquitoes (Diptera: Culicidae) collected in the USA. Bahia Grande virus (BGV) was isolated on 27 occasions from mosquitoes (Aedes, Culex, Anopheles, Psorophora spp.) collected in Texas, Louisiana, New Mexico and North Dakota, between 1972 and 1979 (Kerschner et al., 1986). The Harlingen strain of BGV was isolated from mosquitoes (Aedes salinarius) collected in Texas, in 2001. MSV was isolated from Aedes sp. mosquitoes collected in Colorado, in 1976 (Kerschner et al., 1986). Neutralising antibodies to BGV have been detected in humans, cattle, sheep, wood rats (Neotoma micropus), and opossums (Didelphis marsupialis) (Kerschner et al., 1986). MSV cross-reacts with and is distinguishable from BGV in both complement-fixation and plaque-reduction neutralisation tests (Kerschner et al., 1986).
Species demarcation criteria
Viruses assigned to different species within the genus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in the N proteins; B) minimum amino acid sequence divergence of 10% in their L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) they can be distinguished in virus neutralisation tests; and F) they occupy different ecological niches as evidenced by differences in vertebrate hosts and or arthropod vectors.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Reed Ranch virus* | RRAV |
Virus names and virus abbreviations are not official ICTV designations.

