Family: Rhabdoviridae
Peter J. Walker, Nicolas Bejerman, Kim R. Blasdell, Humberto Debat, Ralf G. Dietzgen, Anthony R. Fooks, Juliana Freitas-Astúa, Kyle Garver, Hideki Kondo, Pedro Luis Ramos-González, Mang Shi, Robert B. Tesh, Noël Tordo, Nikos Vasilakis and Anna E. Whitfield
The citation for this ICTV Report chapter is the summary published as Walker et al. (2022):
ICTV Virus Taxonomy Profile: Rhabdoviridae 2022, Journal of General Virology, 103: 001689.
Corresponding authors: Peter J. Walker ([email protected]); Nicolas Bejerman ([email protected])
Edited by: Jens H. Kuhn, Stuart G. Siddell and Holly R. Hughes
Posted: April 2018, updated November 2019, March 2021, September 2021, January 2023, November 2023, May 2024, October 2024, February 2026
Summary
The family Rhabdoviridae includes four subfamilies, 62 genera and 580 species for viruses with negative-sense, single-stranded RNA genomes of approximately 10–16 kb (Table 1 Rhabdoviridae). Virions are typically enveloped with bullet-shaped or bacilliform morphology but non-enveloped filamentous virions have also been reported. The genomes are usually single RNA molecules with partially complementary termini, but some members have genomes comprising two or three segments. Almost all rhabdovirus genomes have five genes encoding the structural proteins (N, P, M, G and L); however, many rhabdovirus genomes encode other proteins in additional genes or in alternative open reading frames (ORFs) within the structural protein genes, and some rhabdoviruses lack the G gene. The family is ecologically diverse with members infecting plants or animals including mammals, birds, reptiles, amphibians or fish. Rhabdoviruses are also detected in invertebrates, including arthropods, flat worms, round worms or crustaceans. Arthropods may serve as unique hosts or may act as biological vectors for transmission to other animals or plants. Rhabdoviruses include important pathogens of humans, livestock, fish or agricultural crops.
Table 1 Rhabdoviridae. Characteristics of members of the family Rhabdoviridae.
| Characteristic | Description |
| Example | vesicular stomatitis Indiana virus (AF473864), species Vesiculovirus indiana, genus Vesiculovirus |
| Virion | Bullet-shaped or bacilliform particle 100–430 nm in length and 45–100 nm in diameter comprised of a helical nucleocapsid surrounded by a matrix layer and a lipid envelope. Some rhabdoviruses have non-enveloped filamentous virions |
| Genome | Negative-sense, single-stranded RNA of 9.8–16.1 kb (unsegmented, bi-segmented or tri-segmented) |
| Replication | Ribonucleoprotein (RNP) complexes containing anti-genomic RNA are generated and serve as templates for synthesis of nascent RNP complexes containing genomic RNA |
| Translation | From capped and polyadenylated mRNAs transcribed progressively from each gene (3′ to 5′), sometimes containing multiple ORFs |
| Host Range | Vertebrates, invertebrates and plants; many vertebrate and plant rhabdoviruses are arthropod-borne |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, subphylum Haploviricotina, class Monjiviricetes, order Mononegavirales; the family Rhabdoviridae includes 59 genera that are assigned to four subfamilies (Alpharhabdovirinae, Betarhabdovirinae, Gammarhabdovirinae, Deltarhabdovirinae) and three additional unassigned genera (Alphaplatrhavirus, Betaplatrhavirus and Gammaplatrhavirus), together including 580 species. There are at least 300 related, unclassified rhabdoviruses |
Viruses assigned to each of the four subfamilies and 62 genera form a monophyletic clade based on phylogenetic analysis of L sequences. Viruses assigned to each genus usually have a similar genome architecture, including the number and locations of accessory genes, and also have similarities in host range, modes of transmission and/or sites of replication in the cell.
Subfamily Alpharhabdovirinae
The subfamily includes 34 genera for viruses infecting only vertebrates, only invertebrates or vertebrate hosts and arthropod vectors. Viruses assigned to this subfamily sometimes have been referred to informally as dimarhabdoviruses (dipteran and mammalian rhabdoviruses) but this term is misleading as various members may infect birds, reptiles, amphibians, non-dipteran insects, ticks, crustaceans or nematodes.
Genus Almendravirus. The viruses assigned to this genus were isolated from mosquitoes and appear to be poorly adapted (or not adapted) to replication in vertebrates. The genomes of almendraviruses feature an additional gene located between the G and L genes, encoding a small viroporin-like protein.
Genus Alphanemrhavirus. This genus comprises viruses that have been detected by high-throughput sequencing in parasitic nematodes. They are distinct phylogenetically from rhabdoviruses assigned to the genus Betanemrhavirus (subfamily Deltarhabdovirinae). The genomes of alphanemrhaviruses are relatively simple, containing the five structural protein genes, but may include an additional small ORF in the M gene (Mx) overlapping the end of the M ORF. No alphanemrhaviruses have yet been isolated.
Genus Alphapaprhavirus. Alphapaprhaviruses have been detected by high-throughput sequencing in moths and butterflies and are distinct phylogenetically from rhabdoviruses assigned to the genus Betapaprhavirus (subfamily Deltarhabdovirinae). The genomes feature two consecutive genes encoding structurally related class I transmembrane glycoproteins. No alphapaprhaviruses have yet been isolated.
Genus Alpharicinrhavirus. Alpharicinrhaviruses have been detected by high-throughput sequencing in hard ticks and are distinct phylogenetically from rhabdoviruses assigned to the genera Betaricinrhavirus and Gammaricinrhavirus (subfamily Deltarhabdovirinae). The genomes contain only the five canonical rhabdovirus structural protein genes (N, P, M, G and L). One member of the genus appears to lack the G gene. No alpharicinrhaviruses have yet been isolated.
Genus Alphathriprhavirus. Viruses assigned to the genus have been detected in thrips (insects in the family Thripidae) from Italy and China. They are distinct phylogenetically from rhabdoviruses assigned to the genus Betathriprhavirus (subfamily Alpharhabdovirinae). The genomes feature an alternative ORF overlapping the start of the G gene which encodes a class I viroporin-like protein.
Genus Amplylivirus. The viruses assigned to date to the genus Amplylivirus have been identified in amphibians, one being detected in the brain. The genomes contain only the five canonical rhabdovirus structural protein genes (N, P, M, G and L).
Genus Arurhavirus. Arurhaviruses have been isolated from mosquitoes and sandflies. There is evidence of infection in rodents and possibly birds. The genomes feature one or more genes located between the G and L genes, including a gene encoding a viroporin-like protein. An additional gene may also be present between the N and P genes.
Genus Barhavirus. Barhaviruses have been isolated from mosquitoes and infect a range of vertebrates including humans, ruminants, rodents and marsupials. There is no evidence of disease in humans. The genomes are relatively simple but may include a small overlapping ORF encoding a viroporin-like protein at the end of the G gene.
Genus Betathriprhavirus. Viruses assigned to the genus have been detected in thrips (insects in the family Thripidae) from the USA. They are distinct phylogenetically from rhabdoviruses assigned to the genus Alphathriprhavirus (subfamily Alpharhabdovirinae). The genomes feature an additional ORF between the G and L genes which encodes a small protein of unknown function.
Genus Caligrhavirus. Caligrhaviruses have been detected in sea lice (crustaceans) in which they appear to cause active infections. Caligrhavirus genomes are relatively simple, containing the five structural protein genes, but may include an additional gene between the G and L genes. No caligrhavirus has yet been isolated but virions have been observed by electron microscopy.
Genus Cetarhavirus. Cetarhaviruses have been isolated from marine cetaceans (dolphins and porpoises). They are most closely related to perhabdoviruses, siniperhaviruses, scophrhaviruses, all of which infect finfish. The genomes contain only the five canonical rhabdovirus structural protein genes (N, P, M, G and L).
Genus Curiovirus. Curioviruses have been isolated from midges, sandflies and mosquitoes. Vertebrate hosts are largely unknown but there is evidence of infection of birds. The genomes feature one or more genes located between the M and G genes, and one or more genes located between the G and L genes, including a gene encoding a viroporin-like protein.
Genus Ephemerovirus. Viruses assigned to the genus have been isolated primarily from cattle, pigs, mosquitoes or midges, and some members have been detected by metagenomic sequencing of hard ticks. Some ephemeroviruses cause an acute febrile illness in bovines that is seldom fatal. The genomes feature multiple genes between the G and L genes encoding accessory proteins including a non-structural class I transmembrane glycoprotein (GNS) and a viroporin (α1).
Genus Hapavirus. This genus comprises viruses that have been isolated from mosquitoes or midges and that infect birds and mammals. The genomes of hapaviruses are large and complex, featuring multiple accessory genes between P and M genes, and between G and L genes, usually including a gene encoding a viroporin-like protein.
Genus Ledantevirus. Ledanteviruses infect mammals; many have been isolated from bats or rodents and some (or all) may be transmitted by arthropods. Some have been associated with disease in humans or cattle. The genomes are relatively simple but some ledanteviruses feature an additional gene between the G and L genes encoding a small protein of unknown function.
Genus Lostrhavirus. Viruses assigned to the genus Lostrhavirus have been detected in hard ticks, two collected in China and the other taken from an ill human in the USA with a rash. The genome organisations are relatively simple but may include alternative ORFs of unknown importance in the N and P genes.
Genus Lyssavirus. Lyssaviruses circulate among bats (order Chiroptera) and carnivores (order Carnivora) although they may infect all warm-blooded animals causing acute encephalomyelitis with a 99.9% case fatality rate (rabies), once clinical symptoms are observed. Natural transmission is via saliva, usually through a bite by an infected animal. The genomes are relatively simple, containing genes that encode five structural proteins but feature a long 3′-untranslated region (ψ) in the G gene; additional proteins may be expressed from alternative initiation codons in the P gene.
Genus Merhavirus. Merhaviruses have been detected in, or isolated from, culicine mosquitoes. Their genomes contain only the five canonical rhabdovirus structural protein genes (N, P, M, G and L). In one member of the genus, there are two ORFs in the L gene separated by an intron containing eukaryotic spliceosomal intron-splice donor/acceptor sites. This virus replicates in the nucleus and utilizes the host nuclear splicing machinery.
Genus Mousrhavirus. The single virus assigned to date to the genus Mousrhavirus has been isolated on multiple occasions from mosquitoes in Côte d’Ivoire. The genome contains only the five canonical rhabdovirus structural protein genes (N, P, M, G and L).
Genus Ohlsrhavirus. Ohlsrhaviruses have been detected in culicine mosquitoes from Europe, Africa, Asia and the Americas. Although vertebrate hosts of ohlsrhaviruses have not been identified, mosquitoes of the species from which they have been isolated are known viral vectors. Their genomes contain only the five canonical rhabdovirus structural protein genes (N, P, M, G and L).
Genus Perhabdovirus. Perhabdoviruses infect a wide range of teleost fish. They are transmitted through contaminated water and can cause severe haemorrhagic disease. The genomes are relatively simple, containing the five structural protein genes and short intergenic regions. Perhabdoviruses are phylogenetically related to but distinct from fish rhabdoviruses assigned to the genera Sprivivirus, Siniperhavirus and Scophrhavirus.
Genus Replylivirus. The single virus assigned to date to the genus Replylivirus was identified in the brain of a reptile from Cuba. The genome contains the five canonical rhabdovirus structural protein genes (N, P, M, G and L) and a long intergenic region between the G and L genes.
Genus Sawgrhavirus. Sawgrhaviruses have been isolated from hard ticks collected in North America. Their genomes contain the five canonical rhabdovirus structural protein genes (N, P, M, G and L); alternative ORFs of unknown importance are conserved in the G genes of some sawgrhaviruses.
Genus Scophrhavirus. Scophrhaviruses have been isolated from ray-finned fish. They are most closely related to perhabdoviruses and siniperhaviruses, which infect finfish, and cetarhaviruses which infect marine mammals. Their genomes contain only the five canonical rhabdovirus structural protein genes (N, P, M, G and L).
Genus Sigmavirus. Sigmaviruses are transmitted vertically, each virus infecting flies of a single species in the families Drosophilidae, Muscidae, Tephritidae or Hippoboscidae. They have also been detected in honey bees and bird fecal samples by metagenomic sequencing. Infection results in paralysis or death of flies upon exposure to carbon dioxide. The genomes may feature an additional gene (X) located between the M and G genes, encoding a protein of unknown function.
Genus Siniperhavirus. Siniperhaviruses have been isolated from ray-finned fish (class Actinopterygii). They are most closely related to perhabdoviruses and scophrhaviruses, which infect finfish, and cetarhaviruses which infect marine mammals. The genomes contain only the five canonical rhabdovirus structural protein genes (N, P, M, G and L).
Genus Sprivivirus. The viruses assigned to this genus infect a wide range of teleost fish. They are transmitted through contaminated water and can cause severe haemorrhagic disease. The genomes of spriviviruses are relatively simple, containing the five structural protein genes and short intergenic regions. Spriviviruses are phylogenetically related to but distinct from fish rhabdoviruses assigned to the genera Perhabdovirus, Siniperhavirus and Scophrhavirus.
Genus Sripuvirus. Viruses assigned to this genus have been isolated from either sandflies or lizards. The genomes of sripuviruses feature a small protein encoded in a consecutive ORF in the M gene and a small transmembrane protein encoded in an alternative ORF at the start of the G gene.
Genus Sunrhavirus. Sunrhaviruses have been isolated from culicine mosquitoes, biting midges and birds in Africa, Australia and South America. Their genomes feature an additional gene between the M and G genes encoding a small hydrophobic protein and alternative ORFs in several genes including, consistently, the P gene.
Genus Tibrovirus. Some tibroviruses infect cattle and water buffalo and are transmitted by midges; several other tibroviruses have been detected in humans but their role in human disease is unclear. The genomes feature two accessory genes between the M and G genes, and a gene encoding a viroporin-like protein between the G and L genes.
Genus Tupavirus. Tupaviruses have been isolated from birds, insectivores, bats and rodents, and there is evidence of infection in other vertebrates. The genomes feature a long alternative ORF in the P gene and an additional gene encoding a small hydrophobic protein between the M and G genes.
Genus Uniorhavirus. The single virus assigned to the genus has been detected in freshwater mussels in the USA. The genome features two consecutive genes encoding structurally related class I transmembrane glycoproteins.
Genus Vesiculovirus. Vesiculoviruses infect a wide range of vertebrate hosts and are transmitted by insects; some may also be transmitted amongst vertebrates by direct contact. Several vesiculoviruses cause vesicular stomatitis in livestock and/or have been associated with influenza-like illness and encephalitis in humans. The genomes are relatively simple, containing the five structural protein genes and short intergenic regions, but may also include alternative ORFs in the P gene and use of alternative initiation codons in the M gene.
Genus Zarhavirus. The single virus assigned to date to the genus Zarhavirus was isolated from hard ticks collected in Iran. The genome contains only the five canonical rhabdovirus structural protein genes (N, P, M, G and L).
Subfamily Betarhabdovirinae
The subfamily includes 12 genera for viruses infecting plant hosts and, for most of them, arthropod vectors.
Genus Alphacytorhabdovirus. Alphacytorhabdoviruses infect a wide range of monocot and dicot plants and are transmitted by arthropod vectors (aphids) in which they replicate. Alphacytorhabdoviruses have unsegmented genomes and replicate in the cytoplasm of infected plant cells. Alphacytorhabdoviruses form a monophyletic clade within a larger cluster containing the betacytorhabdoviruses and gammacytorhabdoviruses. They feature an additional gene between the P gene and M gene encoding a movement protein. Many alphacytorhabdoviruses also harbor another gene between the G and L genes, which is a viroporin-like protein, as well as an overlapping ORF within the P gene.
Genus Alphagymnorhavirus. Alphagymnorhaviruses infect gymnosperms and their vector is not known. The genomes are unsegmented, encoding five to six genes where the first is the N gene and the last is the L gene; proteins encoded in the intervening genes have distant similarity to those of betagymnorhaviruses and varicosaviruses but their functions are unknown. Alphagymnorhaviruses form a monophyletic clade within a larger cluster containing the betagymnorhaviruses and varicosaviruses.
Genus Alphanucleorhabdovirus. Alphanucleorhabdoviruses infect a wide range of monocot and dicot plants and are transmitted by arthropod vectors (planthoppers, leafhoppers) in which they replicate. Alphanucleorhabdoviruses have unsegmented genomes and replicate in the nuclei of infected plant cells. Alphanucleorhabdoviruses form a monophyletic clade within a larger cluster containing the betanucleorhabdoviruses, gammanucleorhabdoviruses, and dichorhaviruses. They feature an additional gene between the P gene and M gene encoding a movement protein.
Genus Betacytorhabdovirus. Betacytorhabdoviruses infect a wide range of monocot and dicot plants and are transmitted by arthropod vectors (planthoppers, leafhoppers and whiteflies) in which they replicate. Betacytorhabdoviruses have unsegmented genomes and replicate in the cytoplasm of infected plant cells. Betacytorhabdoviruses form a monophyletic clade within a larger cluster containing the alphacytorhabdoviruses and gammacytorhabdoviruses. They feature an additional gene between the P gene and M gene encoding a movement protein. Several betacytorhabdoviruses also harbor other genes after the L gene, between P and M, and G and L, including one encoding a viroporin-like protein.
Genus Betagymnorhavirus. The single virus assigned to the genus was identified in a gymnosperm and its vector is not known. The genome is unsegmented, encoding five genes where the first is the N gene and the last is the L gene; proteins encoded in the intervening genes have distant similarity to those of alphagymnorhaviruses and varicosaviruses but their functions are unknown. Betagymnorhavirus forms a separate branch within a larger cluster containing the alphagymnorhaviruses and varicosaviruses.
Genus Betanucleorhabdovirus. Betanucleorhabdoviruses infect a wide range of dicot plants and some members are transmitted by aphids in which they replicate. Betanucleorhabdoviruses have unsegmented genomes and replicate in the nuclei of infected plant cells. Betanucleorhabdoviruses form a monophyletic clade, being a sister clade to dichorhaviruses. They feature an additional gene between the P and M genes encoding a movement protein.
Genus Deltanucleorhabdovirus. The two viruses assigned to the genus infect dicot plants and their arthropod vector is unknown. Their unsegmented genomes feature one additional gene between the P gene and M gene, encoding a movement protein. Deltanucleorhabdoviruses are likely to replicate in the nuclei of infected plant cells. Deltanucleorhabdoviruses cluster phylogenetically with alphanucleorhabdoviruses, betanucleorhabdoviruses, gammanucleorhabdoviruses and dichorhaviruses; sequence similarities with other betarhabdoviruses are low.
Genus Dichorhavirus. Dichorhaviruses infect plants and are transmitted by Brevipalpus mites. They cause localised lesions on leaves, stems, and fruits of economically important plants such as citrus, coffee and orchids. The genome of dichorhaviruses is bi-segmented: RNA1 contains the N, P, M and G genes, and an additional gene located between the P gene and M gene encoding a putative movement protein; RNA2 contains the L gene. Dichorhaviruses replicate in the nuclei of infected plant cells. Virions formed in plant cells may lack envelopes.
Genus Gammacytorhabdovirus. Gammacytorhabdoviruses infect a wide range of monocot and dicot plants and their vector is unknown. Gammacytorhabdoviruses have unsegmented genomes and replicate in the cytoplasm of infected plant cells. Gammacytorhabdoviruses form a monophyletic clade within a larger cluster containing the alphacytorhabdoviruses and betacytorhabdoviruses. Gammacytorhabdoviruses lack the G gene, while some of them also lack the M gene and others the P3 gene. They feature an additional gene between the P gene and M gene encoding a movement protein and two of them harbor an additional gene between the M and L genes.
Genus Gammanucleorhabdovirus. The two viruses assigned to species in this genus infect monocot plants and are transmitted by leafhoppers in which they replicate. Their unsegmented genomes feature two additional genes between the P gene and M gene, one of which encodes a movement protein. Gammanucleorhabdoviruses replicate in the nuclei of infected plant cells. Gammanucleorhabdoviruses cluster phylogenetically with alphanucleorhabdoviruses, betanucleorhabdoviruses, and dichorhaviruses but with weak bootstrap support; sequence similarities with other betarhabdoviruses are low.
Genus Trirhavirus. Trirhaviruses infect dicot hosts and their vector is not known. The genome of trirhaviruses is tri-segmented; RNA 1 encodes the L protein; RNA 2 contains 4 to 5 ORFs including the coat protein gene; RNA 3 contains 3 to 4 genes including the P, M and G genes. Phylogenetic analysis based on deduced L protein amino acid sequences places all tri-segmented rhabdoviruses into a distinct clade which is basal to alpha-, beta- and gammacytorhabdoviruses.
Genus Varicosavirus. The varicosavirus lettuce big vein-associated virus (LBVaV) is transmitted in soil and zoospores of a chytrid fungus, Olpidium brassicae. The genome of varicosaviruses is bi-segmented: RNA1 encodes the L protein and, in some members, contains a small ORF preceding the L gene; RNA2 contains 3 to 5 ORFs including the coat protein gene. LBVaV virions observed in plant cells are non-enveloped rods resembling intracellular nucleocapsids of other rhabdoviruses.
Subfamily Gammarhabdovirinae
The subfamily includes two genera for viruses infecting finfish or detected in freshwater mussels. Viruses assigned to this subfamily are very distant phylogenetically from fish and mussel rhabdoviruses assigned to the genera Perhabdovirus, Siniperhavirus, Scophrhavirus, Sprivivirus and Uniorhavirus (subfamily Alpharhabdovirinae).
Genus Margarhavirus. The single virus assigned to the genus has been isolated from freshwater mussels in the USA. The genome includes two accessory protein genes (U1 and U2) between the M and G genes, and very long AT-rich untranslated regions.
Genus Novirhabdovirus. Novirhabdoviruses infect teleost fish of numerous species in which they can cause severe haemorrhagic disease. Transmission is waterborne; there is also evidence for egg-associated transmission. The genomes feature an additional gene (NV) that is located between the G and L genes. The NV protein appears to be involved in evasion of the host interferon response.
Subfamily Deltarhabdovirinae
The subfamily includes eleven genera for viruses detected by metagenomic sequencing of invertebrates. No member of the subfamily has yet been isolated.
Genus Alphacrustrhavirus. The viruses in this genus have been detected in marine crustaceans (order Decapoda). The genomes contain only the five canonical rhabdovirus structural protein genes (N, P, M, G and L).
Genus Alphadrosrhavirus. The viruses in this genus have been detected in flies of various species (Diptera: Drosophilidae). The genomes feature an additional gene between the G and L genes in which there are two overlapping ORFs, each of which encodes a small hydrophobic protein with a strongly predicted transmembrane domain.
Genus Alphahymrhavirus. The viruses in this genus have been detected in ants and wasps (order Hymenoptera) and are distinct phylogenetically from rhabdoviruses assigned to the genera Betahymrhavirus and Gammahymrhavirus. The genomes contain only the five canonical rhabdovirus structural protein genes (N, P, M, G and L).
Genus Betahymrhavirus. The viruses in this genus have been detected in wasps (order Hymenoptera) and are distinct phylogenetically from rhabdoviruses assigned to the genera Alphahymrhavirus and Gammahymrhavirus. The genomes feature an additional gene between the M gene and G gene with two overlapping reading frames and a “slippery” sequence in the overlap region that could allow expression of the second ORF by ribosomal frame-shift.
Genus Betanemrhavirus. The viruses in this genus have been detected in parasitic roundworms (Ascaridida: Ascarididae) and are distinct phylogenetically from rhabdoviruses assigned to the genera Alphanemrhavirus (subfamily Alpharhabdovirinae). The genomes feature an additional gene between the P and M genes.
Genus Betapaprhavirus. The viruses in this genus have been detected in moths and butterflies, and are distinct phylogenetically from rhabdoviruses assigned to the genera Alphapaprhavirus (subfamily Alpharhabdovirinae). The genomes feature an additional gene between the G and L genes encoding a small basic protein.
Genus Betaricinrhavirus. The viruses in this genus have been detected in hard ticks (family Ixodidae) and are distinct phylogenetically from rhabdoviruses assigned to the genera Alpharicinrhavirus (subfamily Alpharhabdovirinae) and Gammnaricinrhavirus. The genomes feature an alternative ORF in the N gene, overlapping the end of the N ORF.
Genus Gammahymrhavirus. The viruses in this genus have been detected in bees and wasps, and are distinct phylogenetically from rhabdoviruses assigned to the genera Alphahymrhavirus and Betahymrhavirus. The genomes feature at least one additional gene between the M and G genes, occasionally with overlapping ORFs.
Genus Gammaricinrhavirus. The viruses in this genus have been detected in hard ticks and are distinct phylogenetically from rhabdoviruses assigned to the genera Alpharicinrhavirus (subfamily Alpharhabdovirinae) and Betaricinrhavirus. The genomes feature an additional gene between the M and G genes encoding a protein of approximately 35 kDa.
Genus Primrhavirus. The members of this genus have been detected only in culicine mosquitoes. The genomes contain only the five canonical structural protein genes (N, P, M, G and L).
Genus Stangrhavirus. The members of this genus have been detected only in culicine mosquitoes. The genomes feature an additional gene between the G and L genes encoding a small highly basic protein (6–7 kDa).
Genus not assigned to a subfamily
Within the Rhabdoviridae, three phylogenetically divergent genera do not fall within any of the four existing subfamilies. All viruses assigned to these genera have been detected by metagenomic sequencing of flatworms, fecal samples, barnacles, or vertebrate tissue samples..
Genus Alphaplatrhavirus. The viruses in this genus have all been detected in trematode or cestode worms, fecal samples or vertebrate visceral tissue samples. The genomes feature one or two additional genes between the G and L genes, occasionally with overlapping ORFs and usually including one ORF encoding a predicted class I viroporin.
Genus Betaplatrhavirus. The viruses in this genus have all been detected in trematode or cestode worms, or vertebrate visceral tissue samples. The genomes feature one or two additional genes between the G and L genes, occasionally with overlapping ORFs and usually including one ORF encoding a predicted class I viroporin.
Genus Gammaplatrhavirus. The viruses in this genus have all been detected in trematode or cestode worms, barnacles, or vertebrate visceral tissue samples. The genomes feature one or two additional genes between the G and L genes, occasionally with overlapping ORFs and usually including one ORF encoding a predicted class I viroporin.
Virion
Morphology
Enveloped virions have been reported to be in the range of 100–460 nm in length and 45–100 nm in diameter (Hummeler et al., 1967, Nakai and Howatson 1968, Hummeler and Koprowski 1969, Francki 1973, Knudson 1973) (Figure 1 Rhabdoviridae). The longer forms may represent virions fused end-to-end. Defective-interfering (DI) virus particles are proportionally shorter (Huang et al., 1966). Viruses infecting vertebrates (subfamilies Alpharhabdovirinae and Gammarhabdovirinae) are typically bullet-shaped or cone-shaped; however, some rhabdoviruses infecting animals as well as unsegmented plant rhabdoviruses (subfamily Betarhabdovirinae) appear bacilliform when fixed prior to staining (Kurz et al., 1986, Vasilakis et al., 2013). In unfixed preparations, they may appear bullet-shaped or pleomorphic. The outer surface of virions (except for the quasi-planar end of bullet-shaped viruses) is covered with projections (peplomers) which are 5–10 nm long and about 3 nm in diameter (Hummeler et al., 1967). They consist of trimers of the viral envelope glycoprotein (G). A honeycomb pattern of peplomers is observed on the surface of some viruses. Internally, the nucleocapsid (30–70 nm in diameter) has helical symmetry and appears to have cross-striations (spacing 4.5–5 nm) in negatively-stained and thin-sectioned virions (Simpson and Hauser 1966, Nakai and Howatson 1968, Cartwright et al., 1972). The nucleocapsid consists of a ribonucleoprotein (RNP) complex comprising the genomic RNA and tightly bound nucleoprotein (N) together with an RNA-directed RNA polymerase (L) and polymerase-associated phosphoprotein (P). The RNP complex is active for transcription and replication: the N-RNA template is processed by L, which contains various enzymatic activities, and its cofactor P (Emerson and Wagner 1972, Emerson and Yu 1975). In the cytoplasm, the RNP complex is uncoiled and filamentous, about 700 nm in length and 20 nm in diameter (Sokol et al., 1969). In the virion, the lipid envelope containing G interacts with the coiled RNP complex via the matrix protein (M). Virions reported for some plant rhabdoviruses appear to lack a viral envelope (Dietzgen et al., 2014). Virion morphology for viruses in the subfamily Deltarhabdovirinae or in genus Platrhavirus (not assigned to a subfamily) is currently unknown.
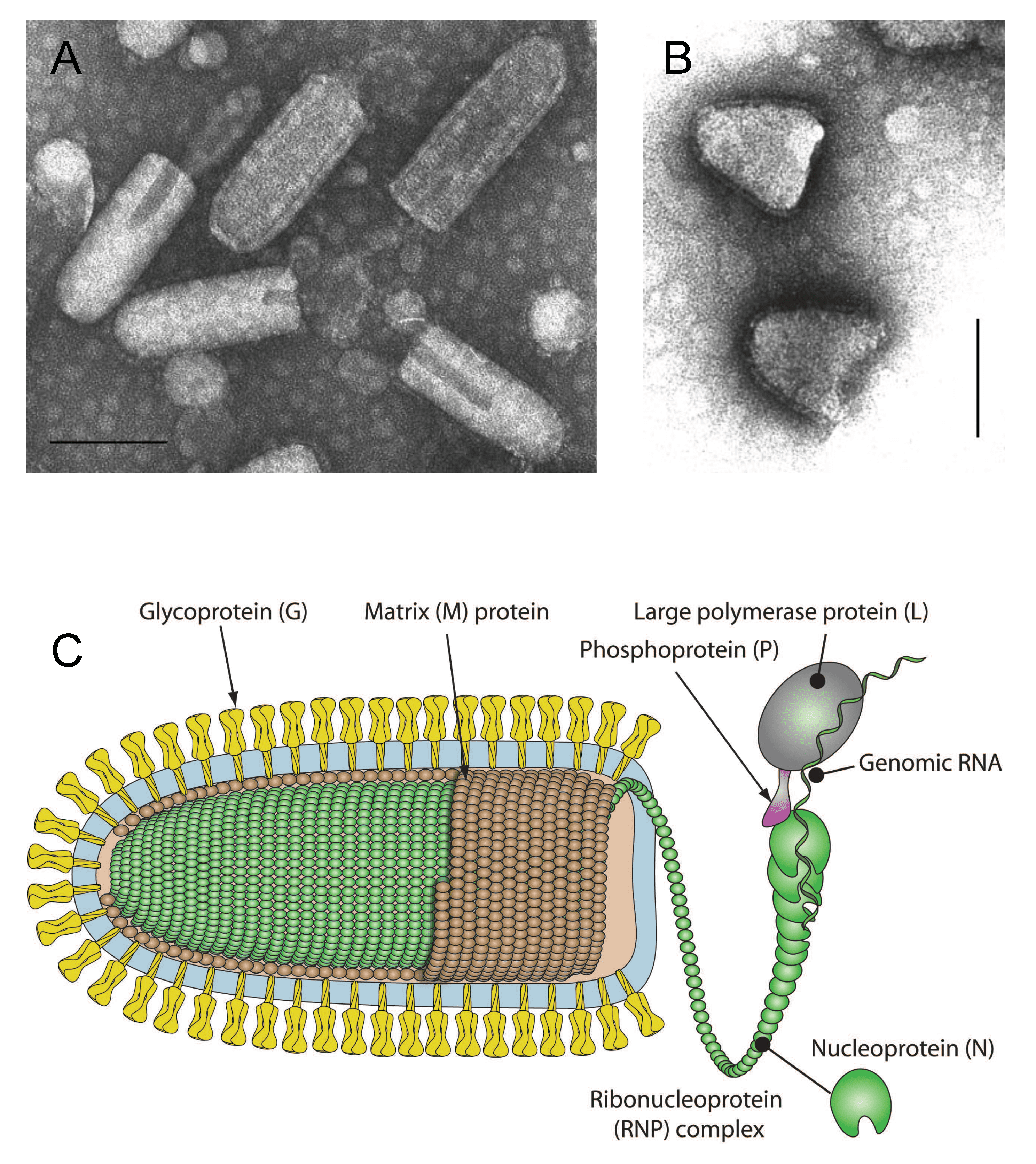 |
| Figure 1 Rhabdoviridae. (A) Negative-contrast electron micrograph of vesicular stomatitis Indiana virus particles. The bar represents 100 nm (Courtesy of P. Perrin). (B) Negative-contrast electron micrograph of rabies virus defective-interfering (DI) particles. (Courtesy of P. Perrin). (C) Schematic illustration of a rhabdovirus virion and ribonucleocapsid structure. Unravelling of the RNP is illustrative only to show more clearly its association with the L and P proteins (Courtesy of P. Le Mercier, ViralZone, SIB Swiss Institute of Bioinformatics). |
Physicochemical and physical properties
Reported virion Mr ranges from 0.3–1.0 × 109 and the S20w is in the range 550–1045 S (plant rhabdoviruses usually have larger S20w values) (Neurath et al., 1966, Jackson and Christie 1977). Virion buoyant density is 1.19–1.20 g cm−3 in CsCl and 1.16–1.19 g cm−3 in sucrose (Warrington 1965, McCombs et al., 1966, Sokol et al., 1968, Jackson and Christie 1977). Virus infectivity is rapidly inactivated at 56 °C, or following UV-, gamma- or X-irradiation, or exposure to formalin or lipid solvents such as detergents (Olitsky and Long 1928, Shechmeister et al., 1962).
Nucleic acid
Virions typically contain a single molecule of linear, negative-sense single-stranded RNA (Mr 3.4 × 106 to 5.4 ×106; approximately 10–16 kb); rhabdoviruses with segmented genomes also may occur with each RNA segment encapsidated independently (Sasaya et al., 2001, Kormelink et al., 2011). The RNA typically represents about 1–3% of virion weight (McSharry and Wagner 1971, Knudson 1973, Thomas et al., 1985). The RNA has a 3′-terminal free hydroxyl group and a 5′-triphosphate and is not polyadenylated (Ehrenfeld and Summers 1972, Moyer et al., 1975). The ends have inverted complementary sequences encoding transcription and replication initiation signals (Keene et al., 1979, Li and Pattnaik 1997, Whelan and Wertz 1999). Defective-interfering RNAs, usually substantially shorter than full-length RNA (less than half length), may be identified in RNA recovered from virus populations (Brown et al., 1967). They are usually negative-sense; however, hairpin RNA forms are also found. Defective-interfering RNAs replicate only in the presence of homologous and, occasionally, certain heterologous helper rhabdoviruses which provide the functional genes (Perrault and Semler 1979, Perrault 1981). Full-length positive-sense RNA, which is an intermediate during the replication process, may constitute a substantial proportion of a viral RNA population (Soria et al., 1974). Like the full-length negative-sense RNA genome, the anti-genome is tightly bound to N and does not occur as naked RNA.
Proteins
Virions generally have five structural proteins (designated N, P, M, G and L; see Table 2 Rhabdoviridae for a summary of their locations, masses and functions). Some rhabdoviruses lack a transmembrane glycoprotein (G). The structural proteins represent 65–75% of dry weight of the virion (McSharry and Wagner 1971, Knudson 1973, Thomas et al., 1985). The function(s) of each of these proteins have been determined largely from studies of the model rhabdoviruses, vesicular stomatitis Indiana virus (VSIV) and/or rabies virus (RABV); the same functions are typically assumed to apply to other rhabdoviruses, although this is not often confirmed experimentally. Most rhabdoviruses also encode multiple additional (accessory) proteins but few of the encoded proteins have been characterised. Ephemeroviruses express a class 1a viroporin (α1) and proteins with viroporin-like structures occur commonly in animal rhabdoviruses (Joubert et al., 2014, Walker et al., 2015) and plant cytorhabdoviruses (genera Alphacytorhabdovirus, Betacytorhabdovirus and Gammacytorhabdovirus). Ephemeroviruses also express large non-structural class I transmembrane glycoproteins (GNS) that is related to the envelope glycoprotein (G) and appears to have arisen by gene duplication (Walker et al., 1992, Wang and Walker 1993, Gubala et al., 2010). Duplication of the G gene also occurs in some hapaviruses (Gubala et al., 2010) as well as alphapaprhaviruses and uniorhaviruses (Longdon et al., 2015, Shi et al., 2016, Goldberg et al., 2023). The function of duplicated G proteins is unknown. Novirhabdoviruses infecting fish express a non-structural protein (NV) that appears to be required for efficient replication and plays a role in evading the host innate anti-viral response (Kurath and Leong 1985, Biacchesi 2011). Plant-adapted viruses have one or more additional non-structural proteins, one of which has been shown to facilitate virus movement between plant cells (Jackson et al., 2005b). Vesiculovirus express two small proteins (C and C′) from an alternative ORF in the P gene (Spiropoulou and Nichol 1993, Peluso et al., 1996); in lyssaviruses, variant forms of P are expressed from alternative initiation codons in the same frame and are involved in modulating the interferon response (Chenik et al., 1995, Moseley et al., 2007).
Alphagymnorhaviruses, betagymnorhavirus and varicosaviruses, besides the N and L proteins, have several proteins (between two and five) that do not resemble the P, M or G proteins usually encoded by rhabdoviruses and lack identified conserved domains (Bejerman et al., 2022). Their functions are unknown. Trirhaviruses, besides the N and L proteins, encode three proteins in their RNA 3 resembling P, M and G proteins, while the functions of the other proteins encoded in their genomes remain unknown (Bejerman et al., 2023).
For certain rhabdoviruses, other nomenclature has previously been used for P (NS, M1 or M2) and M (M1 or M2). The large number and diversity of accessory proteins encoded in rhabdovirus genomes have presented challenges for nomenclature. Some well-described accessory proteins have established names that are in common use. However, as the amino acid sequences of most accessory proteins are not highly conserved and their functions are largely unknown, a universal system of nomenclature based on genome location rather than structural or functional homology has been proposed (Walker et al., 2015). According to this system: i) each additional transcriptional unit (other than N, P, M, G and L) is designated U (unknown) followed by a number in the order they appear in the genome in positive polarity (i.e., U1, U2, U3, etc); ii) the first ORF within each transcriptional unit is assigned the same designation as the transcriptional unit; and iii) each subsequent ORF (alternative, overlapping or consecutive) within any transcriptional unit is designated with a letter (i.e., U1x, U1y, U1z). Alternative ORFs are defined as those which occur within a longer ORF; overlapping ORFs are alternative ORFs that commence within and extend beyond the primary ORF; and consecutive ORFs are those which do not overlap but follow consecutively within the same transcriptional unit. The VSIV C and C′ proteins (55 and 65 amino acids, respectively) are the smallest rhabdovirus proteins known to be expressed in infected cells (Spiropoulou and Nichol 1993, Peluso et al., 1996) and so ORFs ≥ 180 nucleotides may be considered as potentially significant, depending on their location in the transcriptional unit, the Kozak context of the initiation codon and their conservation in multiple virus isolates or related rhabdoviruses (Walker et al., 2015).
Table 2 Rhabdoviridae. Location and functions of rhabdovirus structural proteins.
| Protein | Location, mass and function |
| L | A component of the viral nucleocapsid (ca. 220–240 kDa) responsible for most of the functions required for transcription and replication: RdRP, mRNA 5′-capping, 3′-poly(A) synthesis and protein kinase activities. Observed masses by SDS-PAGE are 150–240 kDa. |
| G | Associates into trimers to form the virus surface peplomers (monomer ca. 65–90 kDa). Binds to host cell receptor(s), induces virus endocytosis then mediates fusion of viral and endosomal membranes. G is variously N-glycosylated and palmitoylated; it lacks O-linked glycans and may have hemagglutinin activity. Induces and binds virus-neutralizing antibodies and elicits cell-mediated immune responses. In some cases, G is involved in tropism and pathogenicity. |
| N | Major component of the viral nucleocapsid (ca. 47–62 kDa). It associates with full-length negative- and positive-sense genomic RNAs, or defective-interfering RNAs, but not mRNAs. N is an active element of the template, presenting the bases to the polymerase. Newly synthesised N probably modulates the balance between genome transcription and replication by influencing the recognition of the transcription signals. N elicits cell-mediated immune responses and humoral antibodies. In some plant rhabdoviruses, N translocates to a sub-nuclear compartment when co-expressed with the cognate P. |
| P | A cofactor of the viral polymerase (ca. 20–30 kDa). It is variously phosphorylated and generally migrates by SDS-PAGE as a protein of about 40–50 kDa, sometimes faster. P is essential for at least two fundamental functions: (i) it mediates the physical link and the correct positioning of L on the N-RNA template; and (ii) it acts as a chaperone during the synthesis of N, by forming N-P complexes that prevent N from self-aggregation and binding to cellular RNA. During the genome replication process, N is then transferred from these N-P complexes to the nascent viral RNA to ensure its specific encapsidation into new RNPs. P elicits cell-mediated immune responses. In several rhabdoviruses P also plays a fundamental role in evading the host innate anti-viral response. |
| M | A basic protein that is an inner component of the virion (ca. 20–30 kDa). It is believed to regulate genome RNA transcription. M binds to nucleocapsids and the cytoplasmic domain of G, thereby facilitating the process of budding. It is sometimes phosphorylated or palmitoylated. M is located in the nucleus and inhibits host cell transcription. It also mediates other pathological effects (cell rounding for VSIV, apoptosis for RABV, intracellular accumulation of the inner nuclear membrane for potato yellow dwarf virus (PYDV). |
Lipids
Based upon studies primarily of vesiculoviruses and lyssaviruses, virions are composed of about 15–25% lipid, with their composition reflecting that of the host cell membrane where virions bud (McSharry and Wagner 1971, Knudson 1973, Thomas et al., 1985). Generally, phospholipids represent about 55–60%, and sterols and glycolipids about 35–40% of the total lipids. G may have covalently associated fatty acids proximal to the lipid envelope (Gaudin et al., 1991, Santiana et al., 2018).
Carbohydrates
Based upon studies primarily of vesiculoviruses, lyssaviruses and ephemeroviruses, virions are composed of about 3% carbohydrate by weight (McSharry and Wagner 1971, Knudson 1973, Thomas et al., 1985). The carbohydrates are present as N-linked glycan chains on G and as glycolipids. Ephemerovirus GNS is also N-glycosylated (Walker et al., 1991). In mammalian cells, the oligosaccharide chains are generally of the complex type; in insect cells they are of non-complex types (Jarvis and Finn 1995). The number and location of N-glycosylation sites vary for G of different rhabdoviruses.
Genome organisation and replication
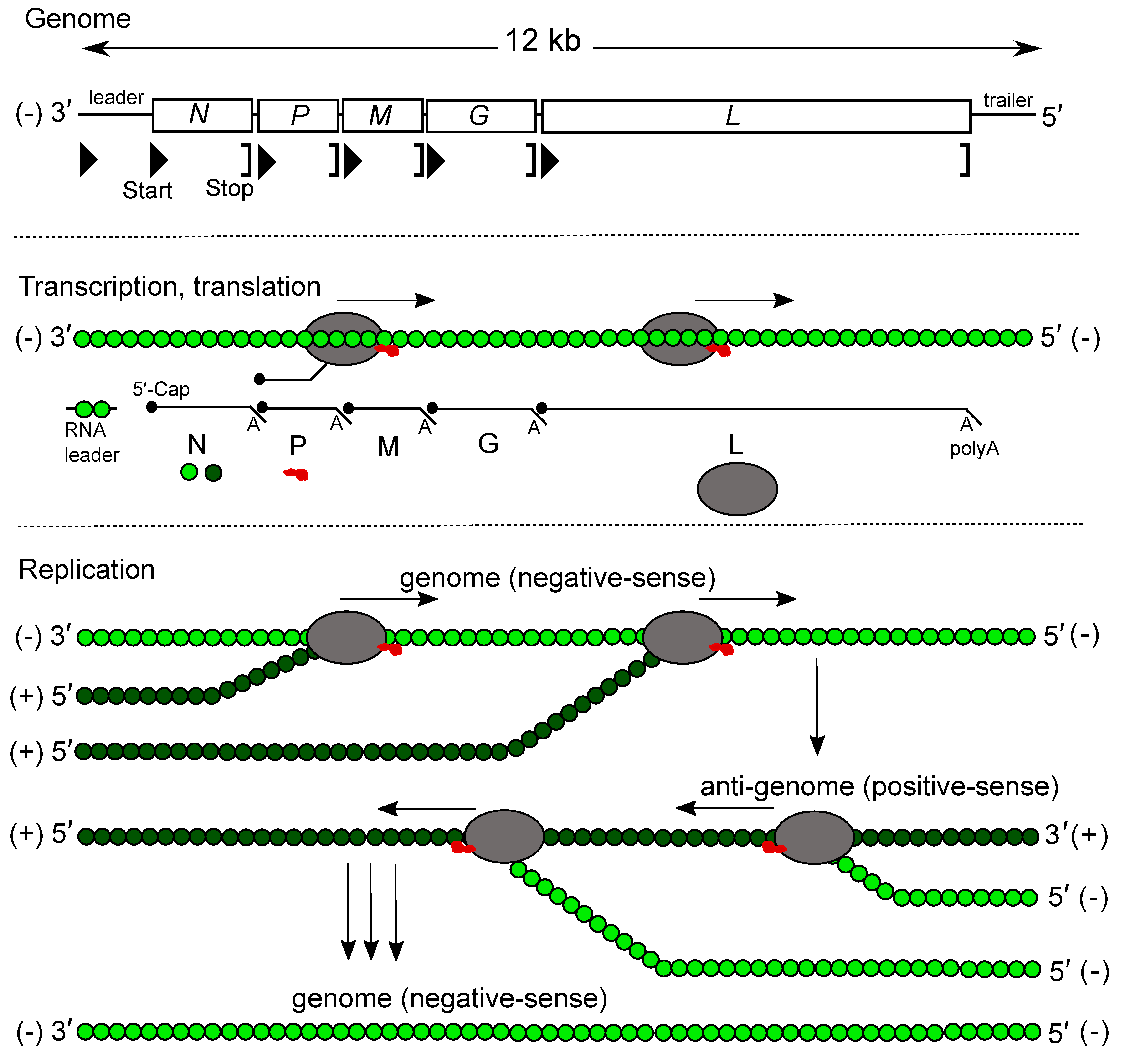
Most rhabdovirus genomes contain at least five ORFs in the negative-sense genome in the order 3′-N-P-M-G-L-5′ (Kuzmin et al., 2009, Walker et al., 2011). The genes are flanked by conserved transcription initiation and termination/polyadenylation signals, about 10 nt in length. Some rhabdoviruses lack the G gene and this could be linked with a transmission mode without the need of a vector (Bejerman et al., 2023). For many rhabdoviruses, additional genes are interposed between the structural protein genes and alternative, overlapping or consecutive ORFs may occur within the structural protein genes or in the additional genes. Some rhabdovirus genomes are segmented. Consequently, genomes of viruses assigned to different genera may vary greatly in length and organisation (Figure 2 Rhabdoviridae).
 |
| Figure 2 Rhabdoviridae. Schematic representation of rhabdovirus genomes shown in reverse (positive-sense) polarity and exemplifying variations in the number and location of accessory genes. A typical member of each genus is represented. Each arrow indicates the position of a long open reading frame (ORF) as described in individual genus sections. ORFs for viroporin-like proteins are coloured yellow and those for movement proteins in blue. Other alternative ORFs occur in some genes; only ORFs (≥180 nt) that appear to have potential to be expressed are shown. |
Most understanding of rhabdovirus replication and transcription has been obtained from studies of vesiculoviruses and lyssaviruses (Banerjee 1987, Banerjee and Barik 1992, Finke and Conzelmann 2005). Genes are transcribed sequentially (from the template virus RNA and in decreasing molar abundance) as 5′-capped, 3′-polyadenylated, monocistronic mRNAs (Ehrenfeld and Summers 1972, Banerjee and Rhodes 1973, Ehrenfeld 1974, Abraham et al., 1975, Moyer et al., 1975, Abraham and Banerjee 1976, Naito and Ishihama 1976) (Figure 3.Rhabdoviridae). A short uncapped, non-polyadenylated and untranslated leader RNA, corresponding to the complement of the 3′-terminus of the viral RNA (i.e., preceding the N mRNA), is also transcribed (Colonno and Banerjee 1976, Colonno and Banerjee 1978). Unlike mRNAs, leader RNA has a 5′-triphosphate terminus (Figure 3 Rhabdoviridae). Leader RNA of some viruses has been identified in the nucleus of infected cells. The mRNAs generally have common 5′-terminal sequences corresponding to the cap structure fused to the first nucleotides copied from the transcription initiation signal. The mRNAs also each contain a 3′-poly(A) tail which is produced by the viral transcriptase upon copying in a reiterative mode at uridine residues present in each transcription termination signal (Naito and Ishihama 1976). Very long 3′-untranslated regions (up to 750 nt) occur in some mRNAs (e.g., lyssaviruses, ephemeroviruses and hapaviruses) (Walker et al., 2015). Intergenic sequences are generally short but may be up to about 100 nt in length. In some cases, the transcription initiation signal of one gene overlaps the 3′-end of the preceding gene.
|
| Figure 3 Rhabdoviridae. Genome organization, transcription and replication of vesicular stomatitis Indiana virus. Top: genome structure. Middle: process of consecutive transcription of leader RNA and messenger RNAs. The role of N (green circles), P (red blob) and L (grey oval) is indicated. Bottom: replication of the negative-sense genome (light green N) via a positive-sense anti-genome intermediate (dark green N). The switch from transcription to replication is regulated by N. The genome and anti-genome strands are not generated in equimolar amounts. |
Non-canonical mechanisms of translation from overlapping or consecutive ORFs appear to occur commonly in viruses assigned to some genera. Although not yet demonstrated experimentally, the likely mechanisms include: i) leaky ribosomal scanning; ii) a stop-start mechanism involving overlapping or consecutive termination and initiation codons and a ‘termination upstream ribosome-binding site’ (TURBS); and iii) ribosomal frame shifts featuring a ‘slippery’ sequence followed by a predicted pseudoknot structure (Walker et al., 2015). In the case of some rhabdoviruses, polycistronic mRNAs result from the read-through of the transcription termination signal, allowing transcription extension across the adjacent 5′-gene. However, in most cases, this appears to be due to corruption of the transcription termination signal during adaptation to growth in cell culture.
Except for plant rhabdoviruses, which generally penetrate plant cells through mechanical damage caused by arthropod or chytrid vectors, rhabdovirus adsorption is mediated by G attachment to cell surface receptors, and penetration of the cell occurs by endocytosis via coated pits (Albertini et al., 2012, Regan and Whittaker 2013). Various candidate receptors have been postulated for RABV (nicotinic acetylcholine receptor AChR, neural cell adhesion molecule NCAM, low affinity nerve growth factor receptor p75NTR), VSIV (phosphatidyl serine), viral hemorrhagic septicemia virus (VHSV) (fibronectin), and others (Schlegel et al., 1983, Bearzotti et al., 1999, Lafon 2005). In addition, carbohydrate moieties, phospholipids and gangliosides may play a complementary role for virus binding (Superti et al., 1984, Superti et al., 1986, Coil and Miller 2004). After penetration by endocytosis, low pH within the endosome triggers fusion between endosomal and viral membranes, liberating the RNP complex into the cytoplasm. The pH-induced fusion depends on conformational changes of the glycoprotein, a process that is reversible upon raising the pH (Roche and Gaudin 2004, Roche et al., 2008). Once the nucleocapsid is released into the cytoplasm, the RNA genome is repetitively transcribed (primary transcription) by the virion transcriptase (Banerjee 1987). N removal does not occur since the transcriptase only recognizes the RNA-N protein complex as template (Arnheiter et al., 1985). The capped and polyadenylated mRNAs are generally translated in cytoplasmic polysomes, except for the G mRNA which is translated on membrane-bound polysomes (Both et al., 1975, Grubman et al., 1975, Morrison and Lodish 1975). Transcription occurs in the presence of protein synthesis inhibitors, indicating that it does not depend on de novo host protein synthesis (Marcus et al., 1971, Villarreal and Holland 1974). Following translation, RNA replication occurs in the cytoplasm (full-length positive-sense and then full-length negative-sense RNA synthesis).
Nucleorhabdoviruses (genera Alphanucleorhabdovirus, Betanucleorhabdovirus, Deltanucleorhabdovirus, Gammanucleorhabdovirus) and dichorhaviruses replicate in viroplasms in the cell nucleus (van Beek et al., 1985, Kitajima et al., 2001, Redinbaugh et al., 2002, Jackson et al., 2005a, Kondo et al., 2013). Replication occurs on the RNA-N protein complex and requires the newly synthesised N, P and L species to concomitantly encapsidate the nascent RNA into a nucleocapsid structure. Apart from freshly translated N, P and L, replication may require host factors. Vesiculoviruses can replicate in enucleated cells, indicating that newly synthesised host gene products are not required (Follett et al., 1974, Wiktor and Koprowski 1974). However, as for some other negative-sense RNA viruses, trafficking of rabies virus proteins to and from the nucleus appears to play an important role in pathogenesis and modulating the host immune response to infection (Wiltzer et al., 2012, Audsley et al., 2016).
It has been proposed that the concomitant binding of N to the nascent positive- or negative-sense viral RNA species may promote replication rather than transcription, by favouring read-through of transcription termination signals (Blumberg et al., 1981, Arnheiter et al., 1985). Replication leads to the synthesis of a full-length positive-sense anti-genome RNA. This, in turn, serves as a replicative intermediate for the synthesis of negative-sense genome RNA for the progeny virions. Following replication, further rounds of transcription (secondary transcription), translation and replication ensue. A typical feature of negative-sense RNA viruses (shared by all members of the order Mononegavirales) is that the RNA genome (or anti-genome) is never “naked” in the cell but is always encapsidated by the nucleoprotein. This RNA-N complex is the true template recognised by the viral polymerase (transcriptase or replicase) (Emerson and Wagner 1972, Moyer et al., 1991).
Post-translational trafficking and modification of G involve translocation across the endoplasmic reticulum membrane, removal of the amino-proximal signal sequence and stepwise glycosylation in compartments of the Golgi apparatus (Rothman and Lodish 1977, Zilberstein et al., 1980). Depending on the cell, G may move to the plasma membrane, particularly to the basolateral surfaces of polarised cells (Fuller et al., 1984, Pfeiffer et al., 1985).
Viral nucleocapsid structures are assembled in association with M and lipid envelopes containing viral G to form virions (Mebatsion et al., 1999). The site of formation of particles depends on the virus and host cell. For vesiculoviruses, lyssaviruses, ephemeroviruses and novirhabdoviruses, nucleocapsids are synthesised in the cytoplasm and virus particles bud from the plasma membrane in most, but not all cells. Some lyssaviruses produce particles that bud predominantly from intracytoplasmic membranes and in some cases prominent virus-specific cytoplasmic inclusion bodies containing N are observed in infected cells (RABV inclusion bodies are called Negri bodies) (Matsumoto 1962, Matsumoto et al., 1974, Manghani et al., 1986, Lahaye et al., 2009). Cytorhabdoviruses (genera Alphacytorhabdovirus, Betacytorhabdovirus and Gammacytorhabdovirus) bud from intracytoplasmic membranes associated with viroplasms; none have been observed to bud from plasma membranes (Jackson et al., 2005a). Nucleorhabdoviruses and dichorhaviruses bud from the inner nuclear membrane and accumulate in the perinuclear space (van Beek et al., 1985, Redinbaugh et al., 2002, Jackson et al., 2005a).
Depending on the virus and host cell type, rhabdovirus infections may inhibit cellular protein synthesis and cause apoptosis by mechanisms that are mediated by M (Weck and Wagner 1979, Koyama 1995, Jackson and Rossiter 1997, Kopecky et al., 2001, Faria et al., 2005, Larrous et al., 2010). Complementation between viral mutants of related viruses may occur (e.g., between vesiculoviruses), but not between viruses assigned to different genera (Pringle et al., 1971, Repik et al., 1976). Complementation has also been reported to occur by re-utilisation of the structural components of UV-irradiated virus (VSIV). Inter-molecular genetic recombination between different virus isolates is very rare, but intra-molecular recombination may occur during the formation of defective-interfering RNAs. Phenotypic mixing occurs between some animal rhabdoviruses and other enveloped animal viruses (e.g., paramyxoviruses, orthomyxoviruses, retroviruses, herpesviruses).
Biology
Rhabdoviruses are ecologically diverse with members infecting plants or animals, including mammals, birds, reptiles, amphibians or fish (Fu 2005, Kuzmin et al., 2009, Dietzgen and Kuzmin 2012, Kuzmin and Walker 2016). Some of the vertebrate rhabdoviruses have a wide experimental host range; rhabdoviruses infecting plants usually have a narrow host range among higher plants. Rhabdoviruses are also detected in invertebrates, including nematodes, platyhelminths, crustaceans, molluscs and arthropods. Some arthropods may serve as biological vectors for transmission to animals or plants. A diverse range of vertebrate and invertebrate cell lines are susceptible to vertebrate rhabdoviruses in vitro.
Rhabdoviruses are not usually transmitted vertically in vertebrates or plants, but transovarial transmission has been documented in insects, and one plant rhabdovirus lacking the G gene has been suggested to be transmitted vertically in seeds (Lee et al., 2022). Sigmaviruses were recognised first as a congenital infection in fruit flies. Vector transmission may involve mosquitoes, sandflies, midges, aphids, whiteflies, leafhoppers, planthoppers, mites or chytrids. Some viruses are transmitted mechanically in sap or from the body fluids of infected hosts. Mechanical transmission of viruses infecting vertebrates may be by contact, aerosol, bite, or venereal. Fish rhabdoviruses can be transmitted by exposure to contaminated water.
Antigenicity
G induces virus-neutralising antibodies which define vertebrate viruses as serotypes and can provide protective immunity. Antigenic cross-reactions in complement-fixation or indirect immunofluorescence tests occur primarily between rhabdoviruses within a genus and involve antigenic determinants located on the N protein. Cross-reactions in indirect immunofluorescence tests have also been detected between some animal rhabdoviruses that are now assigned to different genera (Tesh et al., 1983, Calisher et al., 1989).
Derivation of names
Almendravirus: from Puerto Almendra, near Iquitos in northern Peru from where Puerto Almendras virus, assigned to the species Almendravirus almendras, was first isolated.
Alphacrustrhavirus: from the alpha group of crustacean rhabdoviruses.
Alphacytorhabdovirus: from the alpha group of rhabdoviruses with their cytoplasmic localisation of virus replication complexes (cyto: from Greek kytos, “cell”).
Alphadrosrhavirus: from the alpha group of drosophila rhabdoviruses.
Alphagymnorhavirus: from the alpha group of gymnosperm-associated rhabdoviruses.
Alphahymrhavirus: from the alpha group of hymenopteran rhabdoviruses.
Alphanemrhavirus: from the alpha group of nematode rhabdoviruses.
Alphanucleorhabdovirus: from the alpha group of rhabdoviruses with their nuclear (nucleo: from Latin nux, nucis, “nut”) localisation of virus replication complexes.
Alphapaprhavirus: from the alpha group of lepidopteran (papilionem: from Latin “butterfly”) rhabdoviruses.
Alpharicinrhavirus: from the alpha group of tick (ricinus: from Latin “tick”) rhabdoviruses.
Alphathriprhavirus: from the alpha group of thrips rhabdoviruses.
Amplylivirus: from amphibian lyssa-like virus.
Arurhavirus: derived from Aruac, an ancient native tribe of Trinidadian Americans after which Aruac virus, assigned to the species Arurhavirus aruac, was named, and rhabdovirus.
Barhavirus: from Bahia Grande, a body of water near Brownsville, Texas, where Bahia Grande virus, assigned to the species Barhavirus bahia, was first isolated, and rhabdovirus.
Betacytorhabdovirus: from the from the beta group of rhabdoviruses with their cytoplasmic localisation of virus replication complexes (cyto: from Greek kytos, “cell”).
Betagymnorhavirus: from the beta group of gymnosperm-associated rhabdoviruses.
Betahymrhavirus: from the beta group of hymenopteran rhabdoviruses.
Betanemrhavirus: from the beta group of nematode rhabdoviruses.
Betanucleorhabdovirus: from the beta group of rhabdoviruses with their nuclear (nucleo: from Latin nux, nucis, “nut”) localisation of virus replication complexes.
Betapaprhavirus: from the beta group of lepidopteran (papilionem: from Latin “butterfly”) rhabdoviruses.
Betaricinrhavirus: from the beta group of tick (ricinus: from Latin “tick”) rhabdoviruses.
Betathriprhavirus: from the beta group of thrips rhabdoviruses.
Caligrhavirus: from Caligidae, the family of copepods that includes sea lice, and rhabdovirus.
Cetarhavirus: from cetacean rhabdovirus after the dolphin and porpoise hosts of viruses in the two member species.
Curiovirus: from Curionopolis, a municipality near Serra Norte in northern Brazil from where Curionopolis virus, assigned to the species Curiovirus curionopolis, was first isolated.
Deltanucleorhabdovirus: from the delta group of rhabdoviruses with their nuclear (nucleo: from Latin nux, nucis, “nut”) localisation of virus replication complexes.
Dichorhavirus: from the bi-segmented characteristic of the viral genomes (Dicho: from Greek, meaning “in two, apart or asunder”).
Ephemerovirus: from the virus bovine ephemeral fever virus, assigned to the species Ephemerovirus febris.
Gammacytorhabdovirus: from the gamma group of rhabdoviruses with their cytoplasmic localisation of virus replication complexes (cyto: from Greek kytos, “cell”).
Gammyhymrhavirus: from the gamma group of hymenopteran rhabdoviruses.
Gammanucleorhabdovirus: from the gamma group of rhabdoviruses with their nuclear (nucleo: from Latin nux, nucis, “nut”) localisation of virus replication complexes.
Gammaricinrhavirus: from the gamma group of tick (ricinus: from Latin “tick”) rhabdoviruses.
Hapavirus: from Hart Park serogroup which is the well-established antigenic designation of Flanders virus, Hart Park virus and several other members of the genus.
Ledantevirus: from the name of the medical centre (Hôpital Aristide Le Dantec) in Dakar, Senegal, where the patient from which Le Dantec virus, assigned to the species Ledantevirus ledantec, was first isolated. The medical centre derives its name from the French military doctor, Aristide Auguste Le Dantec, who established the first medical school in Senegal.
Lostrhavirus: from lone star tick (Amblyomma americanum) in which lone star tick rhabdovirus, assigned to the species Lostrhavirus lonestar, was first detected, and rhabdovirus.
Lyssavirus: from Lyssa, the Greek goddess of madness, rage, and frenzy, reflecting the neurological symptoms of rabies virus infection.
Margarhavirus: from the freshwater mussel family (Margaritiferidae), to which the host species (Margaritifera falcata) of the member rhabdovirus is assigned.
Merhavirus: from Merida, the capital of the Mexican state of Yucatan where Merida virus, assigned to the species Merhavirus merida, was first isolated, and rhabdovirus.
Mousrhavirus: derived from Moussa (a coffee plantation in Côte d’Ivoire) where Moussa virus, assigned to the species Mousrhavirus moussa, was first isolated, and rhabdovirus.
Novirhabdovirus: from the additional gene (NV), common to members of the genus and encoding a unique non-virion protein.
Ohlsrhavirus: from Ohlsdorf in Germany, where Ohlsdorf virus, assigned to the species Ohlsrhavirus ohlsdorf, was first detected in mosquitoes, and rhabdovirus.
Perhabdovirus: from perch rhabdovirus, assigned to the species Perhabdovirus perca.
Platrhavirus: from the phylum (Platyhelminthes) of worms in thish the rhabdoviruses have been detected.
Primrhavirus: from Primus virus, a member of one of the species of rhabdoviruses assigned to the genus.
Replylivirus: from reptilian lyssa-like virus.
Rhabdoviridae: from rhabdos (Greek) meaning rod, referring to virion morphology.
Sawgrhavirus: from Sawgrass Lake in Florida where Sawgrass virus, assigned to the species Sawgrhavirus sawgrass, was first isolated, and rhabdovirus.
Scophrhavirus: the genus of the source host (turbot fish; Scophthalmus maximus) from which Scophthalmus maximus rhabdovirus, assigned to the species Scophrhavirus maximus, was first isolated.
Sigmavirus: from the virus discovered in fruit flies (Drosophila melanogaster) that was named virus “sigma” (L'Heritier 1958).
Siniperhavirus: the genus of the source host (mandarin fish; Siniperca chuatsi) from which Siniperca chuatsi rhabdovirus, assigned to the species Siniperhavirus chuatsi, was first isolated.
Sprivivirus: from spring viraemia of carp virus, assigned to the species Sprivivirus cyprinus.
Sripuvirus: from Sripur, the location in north-eastern Bangladesh from where Sripur virus, assigned to the species Sripuvirus sripur, was first isolated.
Stangrhavirus: from Stang virus, a member of one of the species of rhabdoviruses assigned to the genus.
Sunrhavirus: from Sunguru, a village in the Arua District of Uganda from where Sunguru virus, assigned to the species Sunrhavirus sunguru, was first isolated, and rhabdovirus.
Tibrovirus: from Tibrogargan virus, assigned to the species Tibrovirus tibrogargan.
Trirhavirus: from the tri-segmented group of plant rhabdoviruses.
Tupavirus: from the scientific name of the northern tree shrew (Tupaia belangeri) from which tupaia rhabdovirus, assigned to the species Tupavirus tupaia, was first isolated.
Uniorhavirus: from the freshwater mussel family (Unionidae), to which the host species (Lampsilis cardium) of the member rhabdovirus is assigned.
Varicosavirus: from varicose (Latin varix), meaning abnormal dilation or enlargement of a vein or artery, the symptom previously thought to be induced by lettuce big vein-associated virus (LBVaV). However, lettuce big-vein disease, although long thought to be induced by a virus previously designated lettuce big-vein virus, is now considered to be caused by members of the species Ophiovirus mirafioriense (family Aspiviridae, genus Ophiovirus). Viruses of this species are soil-borne and often occur in lettuce together with isolates of LBVaV.
Vesiculovirus: from vesicular stomatitis and the associated lesions that can occur in the mouth and on hooves and udders of animals infected with some vesiculoviruses.
Zarhavirus: from Zahedan, Iran, where Zahedan rhabdovirus, assigned to the species Zarhavirus zahedan, was first isolated, and rhabdovirus.
Subfamily demarcation criteria
Four subfamilies have now been established. Viruses assigned to each subfamily form a monophyletic clade in well supported Maximum Likelihood or Maximum Clade Credibility trees using full-length L sequences. Demarcation of subfamilies is based on significant differences in genome sequence and ecology as indicated by host range and/or the source organisms in which the viruses have been detected.
Genus demarcation criteria
Sixty-two genera have been established to date. Viruses assigned to a genus form a monophyletic clade in well-supported Maximum Likelihood or Maximum Clade Credibility trees using full-length L sequences. The use of L for taxonomic purposes is justified by the presence of broadly conserved domains and the rarity of genetic recombination. Demarcation of genera is based upon considerations of significant differences in genome sequence and architecture, antigenicity and ecological properties (such as host range, pathobiology and transmission patterns).
Relationships within the family
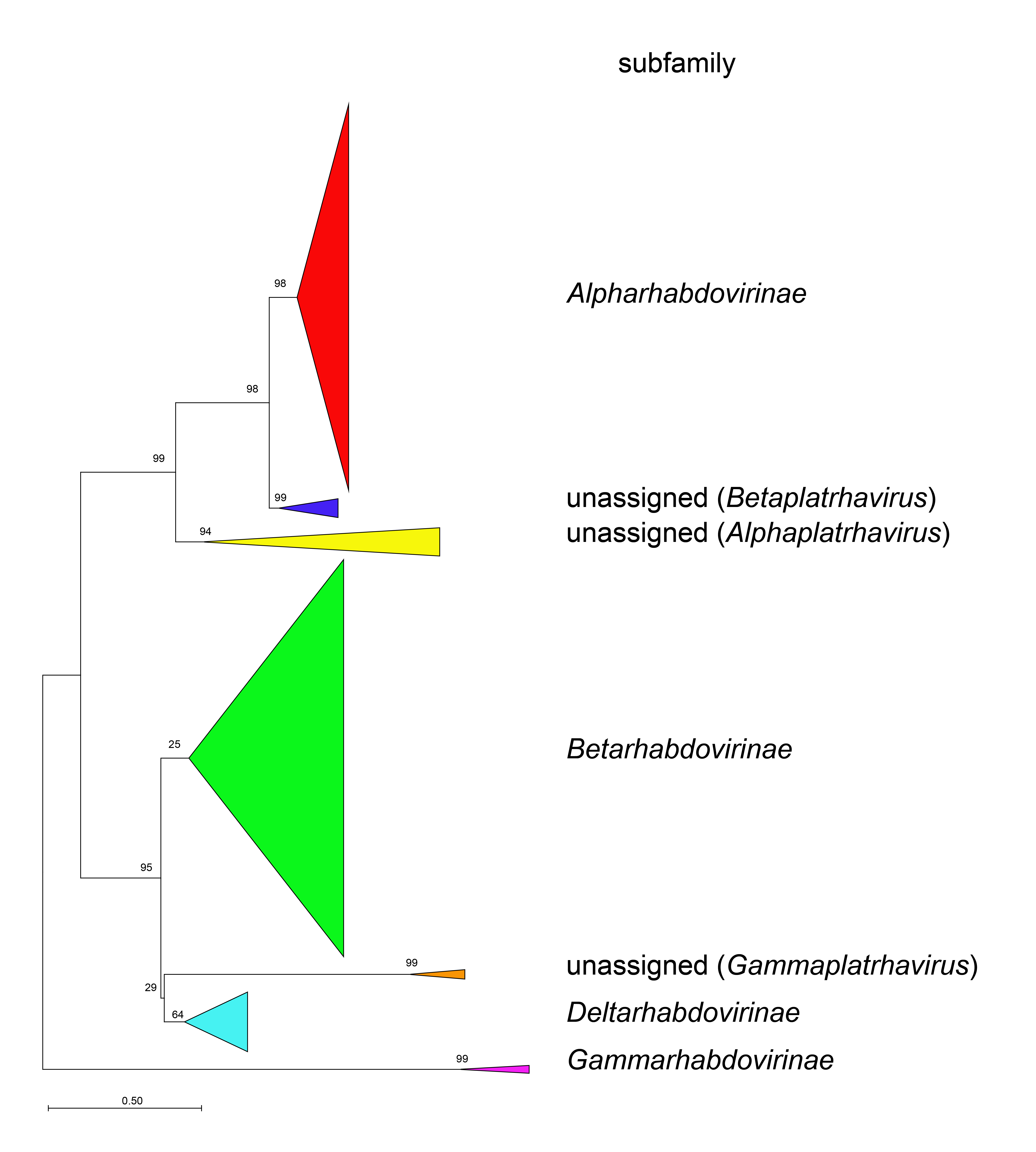
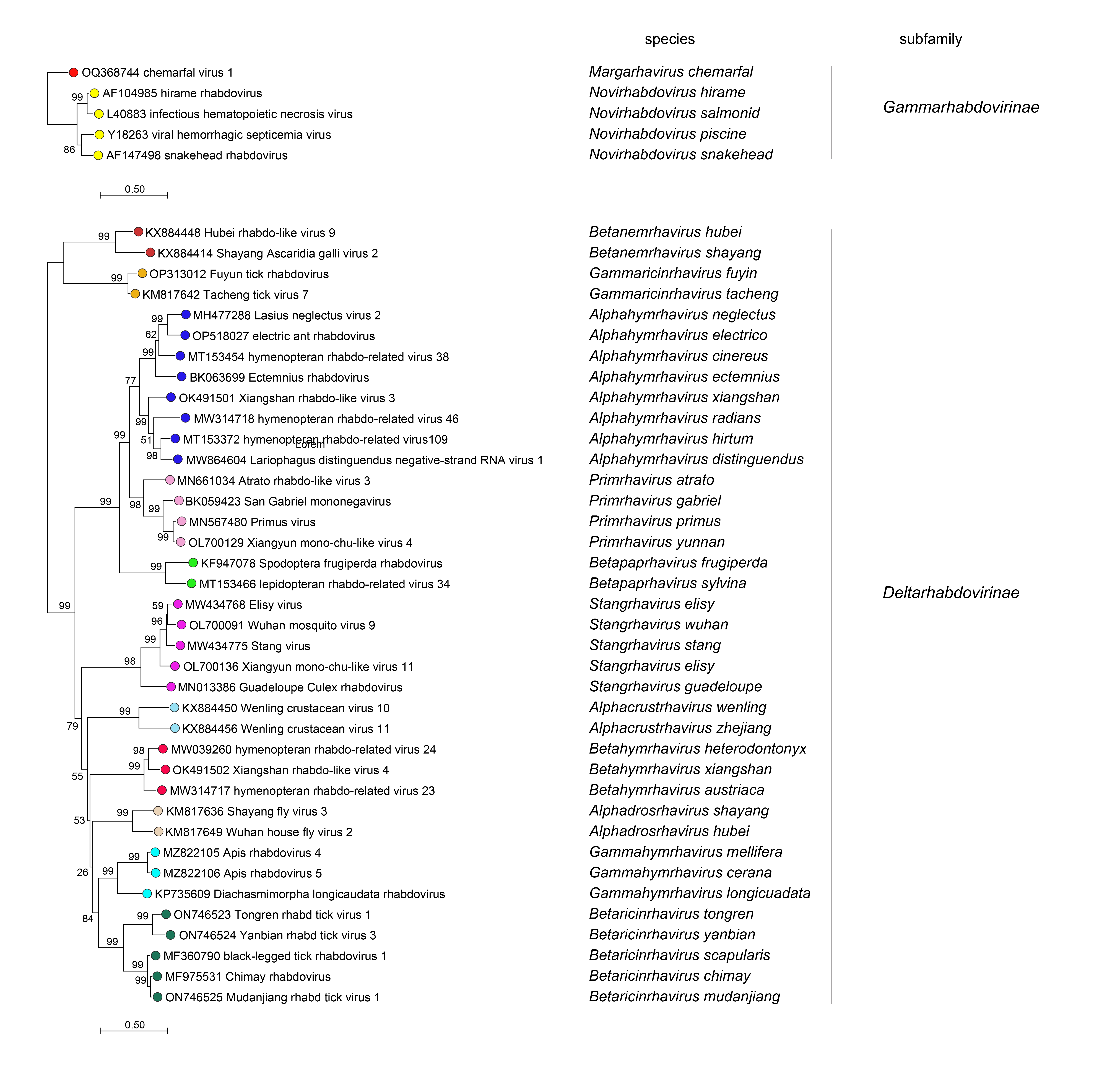
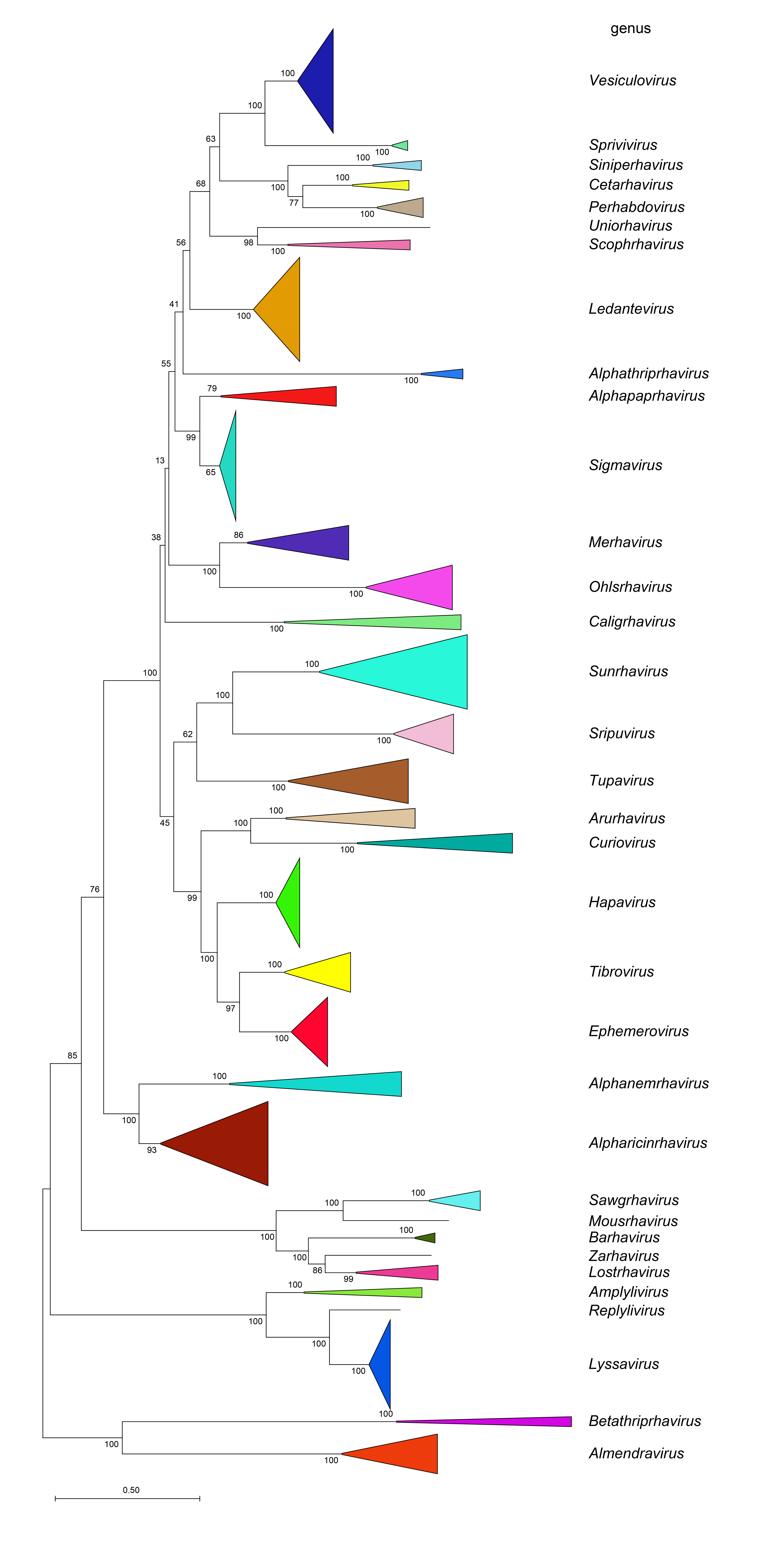
Phylogenetic relationships across the family have been established from Maximum Likelihood or Maximum Clade Credibility trees generated from conserved regions of phylogenetically informative sequence in L (Figure 4 Rhabdoviridae, Figure 5 Rhabdoviridae, Figure 6 Rhabdoviridae). These can be identified by aligning full-length L sequences and eliminating ambiguously aligned regions using the trimAl algorithm (Capella-Gutiérrez et al., 2009). Phylogenetic relationships between viruses assigned to more closely related genera and within genera can also be established using other structural protein genes, notably N and G.
 |
 |
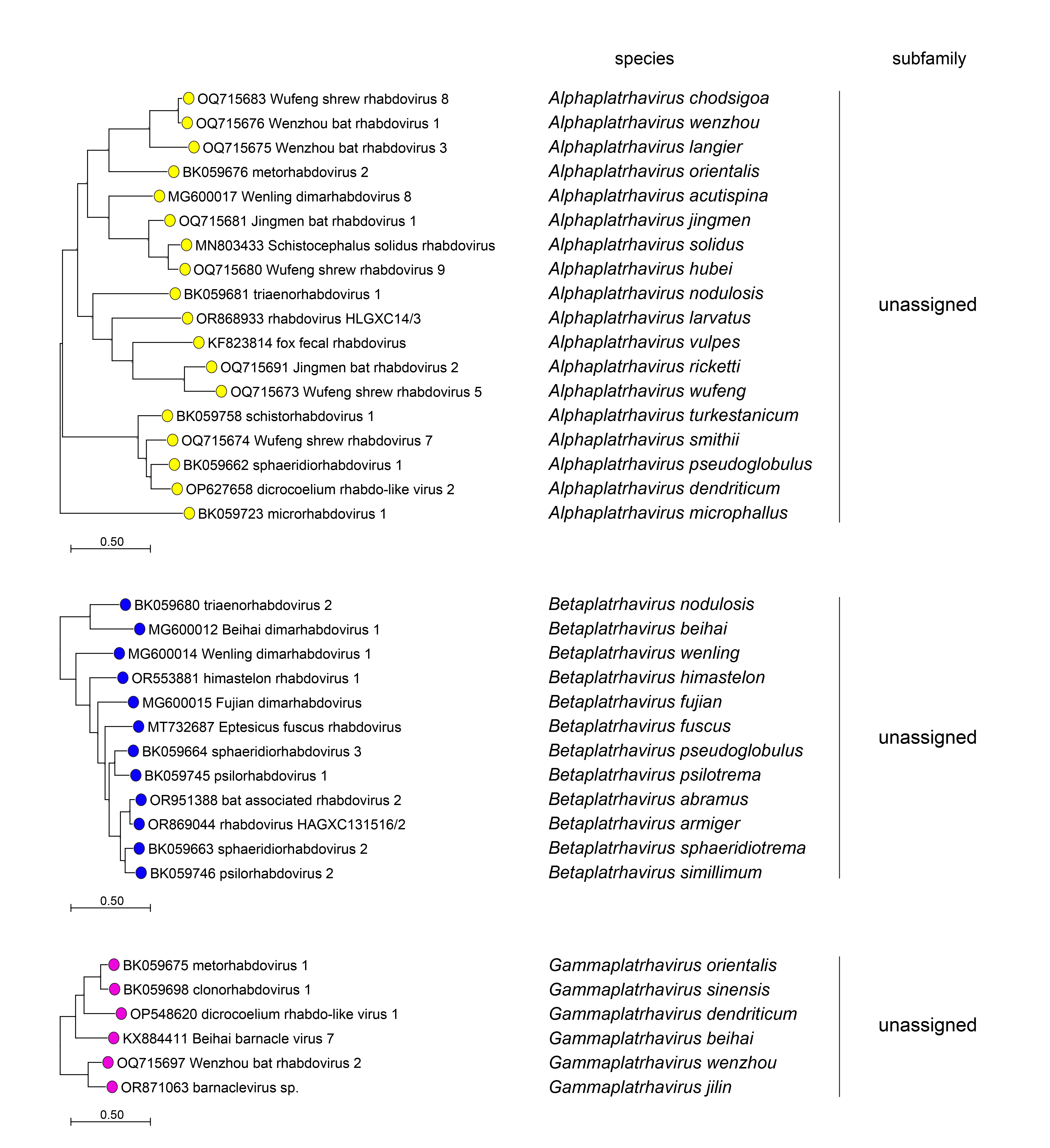 |
| Figure 4 Rhabdoviridae. Top. Maximum-Likelihood (ML) tree inferred from a MAFFT alignment (Katoh and Standley 2013) of full-length rhabdovirus L sequences of 578 rhabdoviruses. Poorly aligned regions were pruned using TrimAl (Capella-Gutiérrez et al., 2009) with 688 positions remaining in the final data set. Clades representing each subfamily and three unassigned genera have been collapsed. Middle. Expanded subtrees selected from the tree with clades expanded to show all members of each genus in the subfamilies Gammarhabdovirinae and Deltarhabdovirinae. Bottom. Expanded subtrees selected from the tree with clades expanded to show all members of the genera Alphaplatrhavirus, Betaplatrhavirus and Gammaplatrhavirus. The evolutionary history was inferred in MEGA7 (Kumar et al., 2016) by using the LG + frequency model of amino acid substitution (Le and Gascuel 2008) with sub-tree pruning and re-grafting (SPR) branch-swapping. The initial tree for the heuristic search was obtained automatically by applying the neighbour-joining algorithm to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree with the highest log likelihood (-327365.75) is shown. Bootstrap proportions following 100 iterations are indicated. The trees is drawn to scale, with branch lengths measured in the number of substitutions per site, the scale bar indicating the value. The trees and corresponding sequence alignments are available to download from the Resources page. |
 |
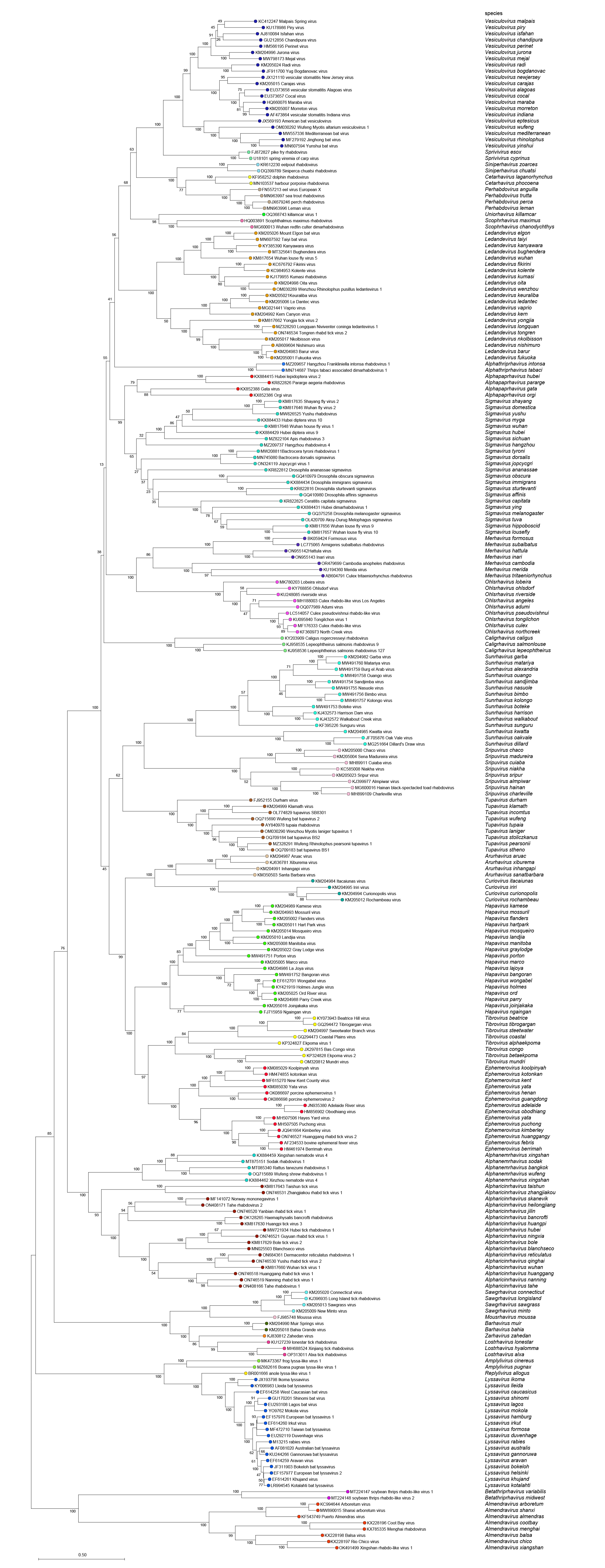 |
| Figure 5 Rhabdoviridae. Top. Maximum-Likelihood (ML) tree inferred from a MAFFT alignment of full-length rhabdovirus L sequences of 246 rhabdoviruses assigned to the subfamily Alpharhabdovirinae. Poorly aligned regions pruned using TrimAl with 1612 positions remaining in the final data set. Clades representing each of the 34 genera have been collapsed. Bottom. The tree expanded with all member viruses shown. The evolutionary history was inferred in MEGA7 by using the WAG + frequency model (Whelan and Goldman 2001) with sub-tree pruning and re-grafting (SPR) branch-swapping. The initial tree for the heuristic search was obtained automatically by applying the neighbour-joining algorithm to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree with the highest log likelihood (-428635.68) is shown. Bootstrap proportions following 100 iterations are indicated. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site, the scale bar indicating the value. The trees and corresponding sequence alignments are available to download from the Resources page. |
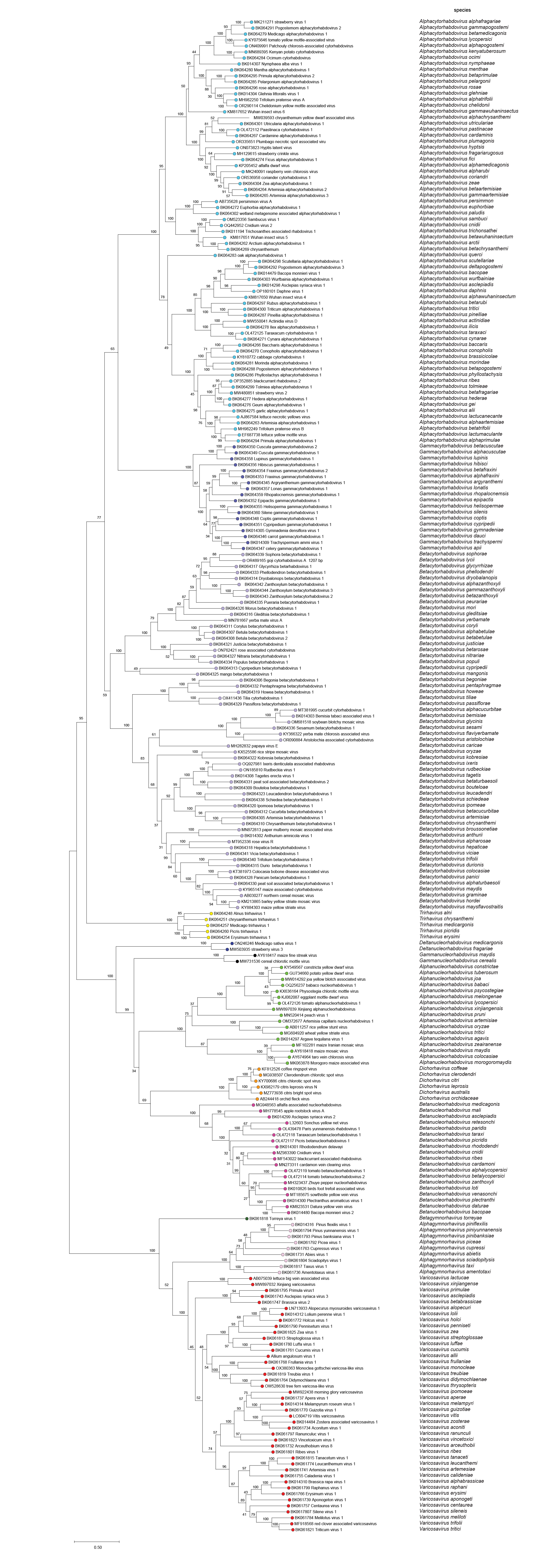 |
| Figure 6 Rhabdoviridae. Maximum-Likelihood (ML) tree inferred from a MAFFT alignment of full-length rhabdovirus L sequences of 253 rhabdoviruses assigned to the subfamily Betarhabdovirinae. Poorly aligned regions pruned using TrimAl with 1207 positions remaining in the final data set. The evolutionary history was inferred in MEGA7 by using the WAG + frequency model of amino acid substitution with sub-tree pruning and re-grafting (SPR) branch-swapping. The initial tree for the heuristic search was obtained automatically by applying the neighbour-joining algorithm to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree with the highest log likelihood (-351109.54) is shown. Bootstrap proportions following 100 iterations are indicated. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site, the scale bar indicating the value. The tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Many general characteristics of rhabdovirus genome structure, organisation, replication and transcription are shared with other members of the order Mononegavirales.
Related, unclassified viruses
Unclassified rhabdoviruses (additional unclassified rhabdoviruses that are probable members of existing genera are listed under individual genus descriptions).
| Virus name | Accession number | Virus abbreviation |
| Agrotis ipsilon virus | MN593030 | AipsV |
| American dog tick rhabdovirus 1 | MF360791 | ADTRV1 |
| Apis rhabdovirus 1 | KY354230 | ApRV1 |
| Apis rhabdovirus 2 | KY354233 | ApRV2 |
| Armigeres rhabdo-like virus 2 | OQ067688 | ArmRLV2 |
| Asiatic toad rhabdo-like virus | OQ599891* | AsTRLV |
| Beaumont virus | KF310911* | BEAUV |
| Beihai rhabdo-like virus 1 | KX884412* | BhRLV1 |
| Beihai rhabdo-like virus 2 | KX884413 | BhRLV2 |
| blattodean rhabdo-related virus 14 | MT153532 | BlRRV14 |
| blue crab virus | BCV | |
| brine shrimp rhabdovirus 1 | OL472789 | BSRV1 |
| Caledonia dog whelk rhabdo-like virus 2 | MF190043* | CDWRLV2 |
| coleopteran rhabdo-related virus 10 | MT153511* | CoRRV10 |
| coleopteran rhabdo-related virus 20 | MT153398 | CoRRV20 |
| coleopteran rhabdo-related virus 28 | MT153411 | CoRRV28 |
| coleopteran rhabdo-related virus 29 | MT153538* | CoRRV29 |
| DakArk 7292 virus | ||
| Dielmo rhabdovirus | TSA#* MG-RAST mgm4604249.3 | DMORV |
| dipteran rhabdo-related virus 19 | TSA# | DiRRV19 |
| dipteran rhabdo-related virus 27 | TSA#* | DiRRV27 |
| dipteran rhabdo-related virus 36 | TSA#* | DiRRV36 |
| drain fly rhabdovirus MG2015 | DFRV | |
| Drosophila busckii rhabdovirus | KR822813 | DBusRV |
| Drosophila subobscura rhabdovirus | KR822817* | DSubRV |
| eastern sea garfish-associated rhabdo-like virus | MH716826* | |
| eel virus B12 | EEVB12 | |
| eel virus C26 | EEVC26 | |
| entamoeba virus | ENTAV | |
| Entomophthora rhabdovirus A | MK231099* | EntRVA |
| Farmington virus | KC602379 | FARV |
| Guiyang nephotettix cincticeps rhabdovirus 1 | MZ209638 | GyNciRV1 |
| Hangzhou nephotettix cincticeps rhabdovirus 1 | MZ209636 | HzNciRV1 |
| Hangzhou rhabdovirus 1 | MZ209651 | HzRV1 |
| Hangzhou rhabdovirus 3 | MZ209687 | HzRV3 |
| Hangzhou rhabdovirus 5 | MZ209738* | HzRV5 |
| Hangzhou tipula scripta rhabdovirus 1 | MZ209762 | HzTsRV1 |
| hemipteran rhabdo-related virus 26 | MT153513* | HeRRV26 |
| hemipteran rhabdo-related virus 30 | MT153400* | HeRRV30 |
| hemipteran rhabdo-related virus 47 | MT153561* | HeRRV47 |
| Huangpi tick virus 3 | KM817630 | HpTV3 |
| Hubei dimarhabdovirus virus 2 | KX884426 | HbDRV2 |
| Hubei dimarhabdovirus virus 3 | KX884423 | HbDRV3 |
| Hubei myriapoda virus 7 | KX884443 | HbMV7 |
| Hubei rhabdo-like virus 1 | KX884422 | HbRLV1 |
| Hubei rhabdo-like virus 2 | KX884447 | HbRLV2 |
| Hubei rhabdo-like virus 5 | KX884446* | HbRLV5 |
| Hubei rhabdo-like virus 6 | KX884421* | HbRLV6 |
| Hubei rhabdo-like virus 8 | KX884420* | HbRLV8 |
| hymenopteran almendra-related virus OKIAV1 | MT153550 | HyARV1 |
| hymenopteran rhabdo-related virus OKIAV8 | MT153471 | HyRRV8 |
| hymenopteran rhabdo-related virus 25 | MW288177* | HyRRV25 |
| hymenopteran rhabdo-related virus 40 | MT153562* | HyRRV40 |
| hymenopteran rhabdo-related virus 45 | MW288210* | HyRRV45 |
| Ixodes ricinus associated rhabdovirus | MT181988 | IricRV |
| Jingmen bat rhabdovirus 3 | OQ715698* | JmBRV3 |
| Jingmen bat rhabdovirus 4 | OQ715705* | JmBRV4 |
| Jingshan fly virus 2 | KM817631* | JsFV2 |
| lepidopteran rhabdo-related virus 3 | MT153436* | LeRRV3 |
| lepidopteran rhabdo-related virus 11 | MW288214* | LeRRV11 |
| Lhasa rhabd tick virus 1 | ON746532 | LsRTV1 |
| Longquan rodent rhabdovirus 1 | OQ715729* | LqRRV1 |
| Longquan rodent rhabdovirus 2 | OQ715723* | LqRRV2 |
| Lye Green virus | KU754522* | LGV |
| mantodean rhabdo-related virus 15 | TSA# | MaRRV15 |
| mecopteran rhabdo-related virus 42 | TSA#* | MeRRV42 |
| midge-associated rhabdo-like virus 2 M2C13 | LR701642* | MaRLV2 |
| mononegavirales sp. | MH560586* | |
| murine feces-associated rhabdovirus | MF175079 | MFARV |
| Myotis pequinius bat rhabdovirus | KJ641709* | MpBRV |
| Nephotettix cincticeps negative-stranded RNA virus 1 | MW084959 | NcNSRV1 |
| neuropteran rhabdo-related virus 31 | MW288224* | NeRRV31 |
| Pteromalus puparum negative-strand RNA virus 1 | KX431032 | PpuNSRV1 |
| Rhabdoviridae sp. isolate SD-JN | OP589942 | |
| rhabdovirus HAGXC131516/4 | OR868925* | |
| rhabdovirus HLNNC15/1 | OR869045* | |
| rhabdovirus PPXSC17/1 | OR868965* | |
| Rhipicephalus associated rhabdo-like virus | MH814974 | RaRLV |
| Rhode Island virus | RHIV | |
| Rio Grande cichlid virus | RGRCV | |
| Rondonia rhabdovirus | MN560631* | RONRV |
| Sanya conocephalus maculatus rhabdovirus 1 | MZ209844 | SyCmaRV1 |
| Sanya rhabdovirus 1 | MZ209928* | SyRV1 |
| Shuangao insect virus 6 | KM817639* | SgIV6 |
| sorghum stunt mosaic virus | SSMV | |
| soybean cyst nematode-associated northern cereal mosaic virus | HM849039 | SNaRV |
| Taastrup virus | AY423355* | TAAV |
| Tongliao rhabd tick virus 1 | ON746529 | TlRTV1 |
| ulcerative disease rhabdovirus | UDRV | |
| Varroa jacobsoni rhabdovirus 1 | MT482464 | VjacRV1 |
| Wenling dimarhabdovirus 10 | MG600019* | WlDRV10 |
| Wenzhou bat rhabdovirus 4 | OQ715709* | WzBRV4 |
| Wenzhou bat rhabdovirus 5 | OQ715711* | WzBRV5 |
| Wenzhou rodent rhabdovirus 1 | OQ715685 | WzRRV1 |
| Withyham virus | KU754523* | WITHV |
| Wufeng shrew rhabdovirus 2 | OQ715686 | WfSRV2 |
| Wufeng shrew rhabdovirus 3 | OQ715687 | WfSRV3 |
| Wufeng shrew rhabdovirus 4 | OQ715703* | WfSRV4 |
| Wufeng shrew rhabdovirus 6 | OQ715684 | WfSRV6 |
| Wufeng shrew rhabdovirus 10 | OQ715679 | WfSRV10 |
| Wufeng shrew rhabdovirus 15 | OQ715713* | WfSRV15 |
| Wufeng shrew rhabdovirus 18 | OQ715707* | WfSRV18 |
| Wuhan insect virus 7 | KM817653 | WhIV7 |
| Xiangyun mono-chu-like virus 9 | OL700134* | XyMCLV9 |
| Xiangyun mono-chu-like virus 10 | OL700135 | XyMCLV10 |
| Xiangshan rhabdo-like virus 2 | OK491500 | XsRLV2 |
| Xiangshan rhabdo-like virus 5 | OK491503 | XsRLV5 |
Virus names and virus abbreviations are not official ICTV designations.
* Coding region sequence incomplete
# TSA – transcriptome shotgun assembly. For viruses with OKIAV in their name see doi.org/10.5061/dryad.87vt6hm and (Käfer et al., 2019). For Deilmo rhabdovirus see (Temmam et al., 2016)