Family: Spinareoviridae
Genus: Oryzavirus
Distinguishing features
The oryzavirus genome consists of 10 segments of linear dsRNA. The viruses assigned to species in this genus are rice ragged stunt virus (RRSV) and Echinochloa ragged stunt virus (ERSV) (Chen et al., 1989). The genome of RRSV encodes at least seven putative structural proteins and three nonstructural proteins. Oryzaviruses are turreted reoviruses with distinctive pentameric turrets that sit on the outside of the innermost capsid at each fivefold axis (Miyazaki et al., 2008). Oryzaviruses replicate in delphacid planthoppers but can not be vertically transmitted.
Virion
Morphology
Intact RRSV virions appear to be icosahedral in symmetry and double-layered. The particle diameter is 75–80 nm and surface A-spikes (approximately 10–12 nm wide and 8 nm in length) are attached to the end of B-spikes situated at the five-fold axes of the viral core. The subviral or core particles have an estimated diameter of 57–65 nm (Figure 1 Oryzavirus) and possess 12 B-type spikes, 8–10 nm in height, 23–26 nm wide at the base and 14–17 nm wide at the top. In negatively-stained preparations of RRSV, B-spiked subviral particles have been seen but intact double-layered particles are not seen without pretreatment with fixative. ERSV virions are slightly larger than RRSV particles.
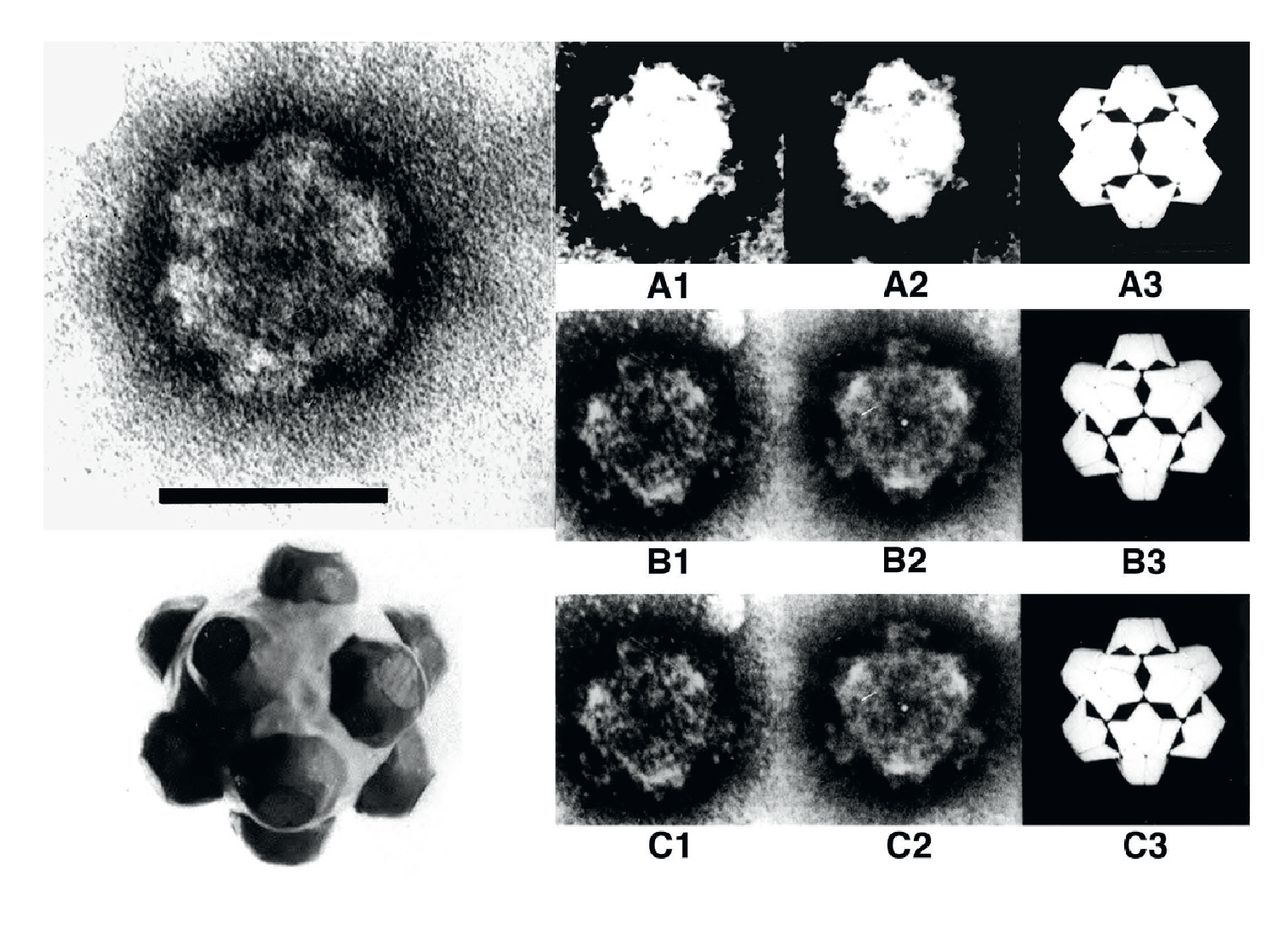 |
| Figure 1 Oryzavirus (Top left) Electron micrograph of rice ragged stunt virions (courtesy of R.G. Milne). (Bottom left) schematic of virion; (right panel) micrographs of the virion having 2-, 3- and 5-fold symmetries (A1, B1 and C1, respectively) images of the same rotated by increments of 180° (A2), or 120° (B2), or 72° (C2) and proposed models of the 2-, 3- and 5-fold symmetries (A3, B3 and C3 respectively) (courtesy of E. Shikata). The bar represents 50 nm. |
Physicochemical and physical properties
RRSV particles sediment as a single component and are stable at pH 6.0–9.0. They are stable in 0.1 M MgCl2. The B spikes dissociate from the core particle in 0.5 M MgCl2 and the entire particle is disrupted in 2 M MgCl2. The particles retain infectivity after 7 days at 4 °C and after 10 min at 50 °C but lose their infectivity after 10 min at 60 °C. They retain infectivity after three cycles of freezing and thawing. The particles contain an RNA-directed RNA polymerase (RdRP).
Nucleic acid
The oryzavirus genome consists of 10 linear dsRNA segments. The genomes of RRSV and ERSV have similar lengths and segment profiles (RRSV Mr 18.15×106 (26,066 bp); ERSV Mr 17.78×106), with segments ranging in length from 1,162 to 3,849 bp. The genomic dsRNAs are termed Seg1 to Seg10, in order of increasing electrophoretic mobility in 7.5% polyacrylamide gels. The entire genome of RRSV has been sequenced; Seg4 and Seg10 are longer than they appear from migration in polyacrylamide gels, suggesting that they may migrate in the position 3 and 9 respectively during agarose gel electrophoresis (AGE). The conserved terminal sequences of the ERSV genome segments are identical to those of RRSV (5′-GAUAAA…(G)GUGC-3′) and differ from those of phytoreoviruses or fijiviruses. RRSV RNAs hybridize weakly with their counterparts in ERSV but not with segments of the phytoreovirus rice dwarf virus (RDV).
Proteins
RRSV particles are composed of five major, highly immunoreactive structural proteins (with estimated masses of 33, 39, 43, 70 and 120 kDa), and at least five minor structural proteins (49, 60, 76, 90 and 94 kDa) (Hagiwara et al., 1986). RRSV RNA segments S5, S8 and S9, respectively, encode a 90 kDa minor structural protein, a 67 kDa major structural protein, which is further self-processed to 46, 43 and 26 kDa proteins, and a 38 kDa major structural protein. P9 is thought to be involved in vector transmission. RRSV S4 probably encodes an RdRP and a second protein of unknown function. ERSV particles have four major structural proteins (127, 123, 63 and 34 kDa) and three minor proteins (103, 50 and 49 kDa). The reported differences in morphology of the outer capsids of RRSV and ERSV could be at least partially due to differences in the masses of these structural proteins. The genome of RRSV encodes at least seven putative structural proteins (P1, P2, P3, P4A, P5, P8B, and P9) and three nonstructural proteins (Pns6, Pns7, and Pns10). Among the structural proteins encoded by RRSV, P2, P3, P4A and P5 are a putative guanylyltransferase, capsid shell protein, RdRP and capping enzyme, respectively. The core particles of RRSV contain core capsid protein P3, P4A protein, and 10 segments of dsRNAs. The major outer capsid protein P8 and spike protein P9 are added onto the core particles to assemble double-layered particles.
Lipids
None reported.
Carbohydrates
There is no evidence for the glycosylation of oryzavirus proteins.
Genome organization and replication
The genome organization is well characterized only for RRSV (Table 1 Oryzavirus). Each of the dsRNA genome segments has a single large ORFs except S4, which has two large ORFs. The proteins encoded by S3, S8 and S9 are major components of the RRSV particle, but those encoded by segments S6, S7 and S10 are not found in the virion. S8 codes for a polyprotein that appears to autocatalytically cleave into at least two polypeptides one of which is a major structural protein. The larger protein encoded by S4 appears to be an RdRP. Among the non-structural proteins encoded by RRSV, Pns6 functions as a viral RNA-silencing suppressor and a viral movement protein (Wu et al., 2010), whilst Pns7 has been identified as an NTP-binding protein. Pns6 and Pns10 are also the components of the matrix of viroplasm to support viral replication and assembly of progeny virons in virus-infected plant hosts or insect vectors (Chen et al., 2014).
Table 1 Oryzavirus Genome segments and protein products of rice ragged stunt virus
| Genome segment | bp | Protein nomenclature | Protein Mr predicted (kDa) | Protein Mr apparent (kDa) | Function (location) |
|---|---|---|---|---|---|
| Seg1 | 3849 | P1 | 137.7 | 137 | Virus core associated, B spike |
| Seg2 | 3810 | P2 | 133.1 | 118 | Guanylyltransferase, inner core capsid |
| Seg3 | 3699 | P3 | 130.8 | 130 | Major core capsid |
| Seg4 | 3823 | P4A (Pol) | 141.4 | 145 | RdRP |
| P4B | 36.9 | Unknown | |||
| Seg5 | 2682 | P5 (Cap) | 91.4 | 90 | Capping enzyme |
| Seg6 | 2157 | Pns6 | 65.6 | Nonstructural protein, viroplasm matrix protein, RNA-silencing suppressor, cell-to-cell movement protein | |
| Seg7 | 1938 | Pns7 | 68 | 66 | Nonstructural protein |
| Seg8 | 1814 | P8 | 67.3 | 67 | Precursor protease, major outer capsid protein |
| P8A/ P8B | 25.6/41.7 | 47/44 | |||
| Seg9 | 1132 | P9 | 38.6 | 37 | Vector transmission, spike |
| Seg10 | 1162 | Pns10 | 32.3 | 32 | Nonstructural protein, viroplasm matrix protein |
Biology
Oryzaviruses infect plants in the family Gramineae, causing diseases in rice (RRSV) and barnyard grasses and millets (Echinochloa spp.; ERSV). They are transmitted by, and replicate in, phloem-feeding, viruliferous delphacid planthoppers (RRSV: Nilaparvata lugens (Stål, 1854); ERSV: Sogatella vibix (Haupt, 1927)). RRSV is ingested when a hopper feeds on rice plants, usually at the seedling stage. The minimum acquisition access period for the vector is about 3 hours, the latent period is about 9 days, and the minimum inoculation access time is about 1 hour. Planthopper nymphs are more efficient vectors than adults, but all forms of the insect can act as vectors. Any individual viruliferous hopper gives intermittent transmission. RRSV is not passed though the egg. After acquisition from rice plants, RRSV is first detected in the epithelial cells of the midgut, from where it proceedes to the visceral muscles surrounding the midgut, then throughout the visceral muscles of the midgut and hindgut, and finally into the salivary glands.
Oryzaviruses appear to replicate in electron-dense viroplasms within the cytoplasm of phloem, or phloem-associated plant cells and in cells of the salivary glands, fat body, gut and brain of the planthopper. The phloem cells proliferate to form galls on the plant. RRSV has been reported in southeastern and far-eastern Asian countries, where it affects rice yields (generally 10–20% loss, but up to 100% in severely affected areas). ERSV has been reported in Taiwan.
Antigenicity
RRSV and ERSV cross-react in serological tests. Polyclonal antisera raised against RRSV particle preparations react most strongly with P3, P8 and P9 (both the native state and the state resulting from in vitro production), suggesting that they are highly immunogenic. P5 is weakly immunogenic. Glutathione-S-transferase-Pns7 fusion protein is highly immunogenic, and antibodies against this protein are useful in ELISA for the detection of RRSV in infected plants and insects.
Species demarcation criteria
In addition to the general criteria used throughout the family, members of different species in the genus Oryzavirus differ in the planthopper vector used and the plant host species.

