Family: Spinareoviridae
Genus: Orthoreovirus
Distinguishing features
Members of the genus Orthoreovirus, colloquially referred to as orthoreoviruses, infect only vertebrates and are spread by respiratory or fecal-oral routes. All members of the genus have a well-defined capsid structure with 12 spikes or turrets situated on the surface of the core particle at the icosahedral vertices. Virons contain 10 linear dsRNA molecules of which three are long (L), three are medium (M), and four are short (S). Orthoreoviruses have a characteristic protein profile with three λ, three µ, and four σ primary translation products and additional short gene products that are encoded by polygenic segments. The genus can be divided functionally into two subgroups based on the ability to induce cell-cell fusion; members of all species, except Orthoreovirus mammalis and Orthoreovirus piscis, encode fusion-associated small transmembrane (FAST) proteins responsible for inducing cell-cell fusion and syncytium formation. Along with the fusogenic aquareoviruses (that encode distinct FAST proteins) and a recently identified rotavirus B isolate (Diller et al., 2019), these are the only well-characterized examples of nonenveloped viruses that are fusogenic, and FAST proteins are the only known examples of membrane fusion proteins encoded by nonenveloped viruses (Ciechonska and Duncan 2014).
Virion
Morphology
Virions are icosahedral with a roughly spherical appearance and a double-layered protein capsid, the different layers of which are discernible by negative-staining and electron microscopy (Figure 1 Orthoreovirus). Higher-resolution images have been obtained by cryo-electron microscopy (cryoEM) and image reconstruction of virus particles from three species, Orthoreovirus mammalis, Orthoreovirus avis, and Orthoreovirus papionis. These particles are similar with a central compartment (about 48 nm in diameter) containing the dsRNA genome segments, surrounded by an inner capsid (60 nm diameter composed of 120 copies of protein λ1) and an outer capsid (85 nm diameter) that has T=13 symmetry. The inner capsid of the orthoreoviruses is equivalent to the T=1 core particle of the rotaviruses and the sub-core of the orbiviruses (which is composed of 120 molecules of VP3 interpreted as having T=2 pseudo icosahedral symmetry). The surface of the complete orthoreovirus particle is covered by 600 finger-like projections arranged in 60 hexameric and 60 tetrameric clusters that surround solvent channels. These channels extend radially into the outer capsid layer.
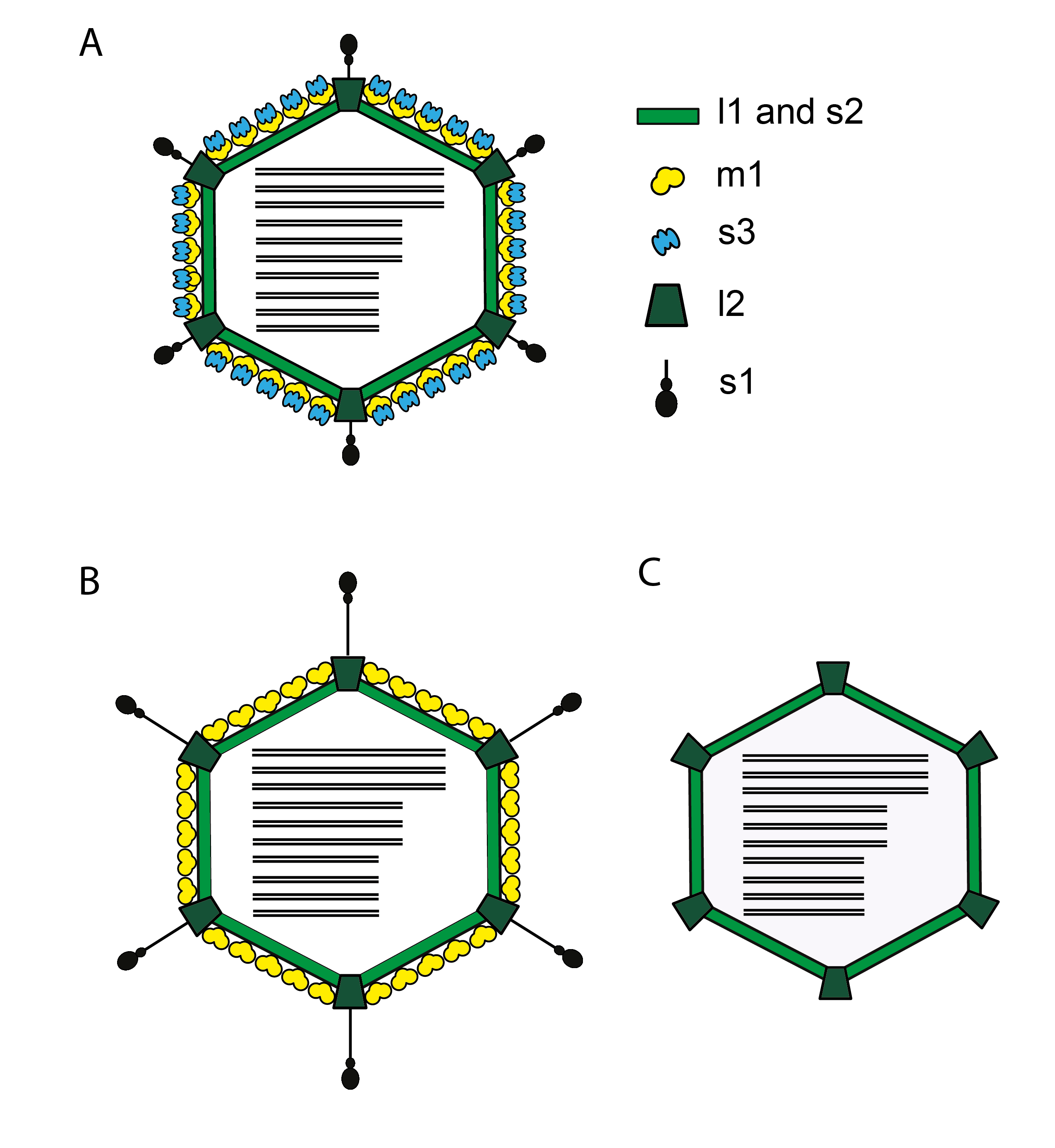 |
| Figure 1 Orthoreovirus Diagrammatic representation of cross-section of an orthoreovirus virion (A) is shown along with the major protein components of the inner (s2, l1) and outer capsids (s1, s3, m1, l2). Schematic representations of the ISVP (B) and the core (C) are shown using the same color scheme. Figure adapted from (Gummersheimer et al., 2021). |
Intact virions from isolates of Orthoreovirus mammalis (collectively referred to as mammalian reoviruses [MRV]) also contain large, open depressions with a flower-shaped structure at the five-fold axes, resulting in an angular capsid profile when viewed in the three-fold orientation. Intermediate subvirion particles (ISVPs), which are generated by partial removal of the outer-capsid proteins, are approximately 80 nm in diameter. The flower-shaped structures at the five-fold axes of the ISVPs may contain an extended form of the viral attachment protein, σ1, which protrudes as a 40 nm fiber from the vertices. Isolates of the species Orthoreovirus piscis and baboon reovirus (BRV, species Orthoreovirus papionis), Mahlapitsi reovirus (MAHLV, species Orthoreovirus mahlapitsiense) and Broome reovirus (BrRV, species Orthoreovirus broomense) do not encode a fiber homologue; the attachment proteins of these viruses have not been identified (Yan et al., 2011). MRV core particles generated by more extensive removal of the outer capsid proteins have been examined by X-ray crystallography and have 150 ellipsoidal nodules (protein σ2) on their surface and distinctive turrets located at the five-fold axes. These projections, which are altered conformations of the flower-shaped structures observed on intact virions (composed of protein λ2, the viral capping enzyme) are about 10 nm in length, possessing central channels 5–8 nm in diameter extending into the central compartment.
Physicochemical and physical properties
The virion Mr is about 130×107 with a buoyant density in CsCl of 1.36 g cm−3 (1.38 g cm−3 for ISVPs, 1.43 g cm−3 for core particles). The virion, ISVP, and core S20,w values are about 730 S, 630 S, and 470 S, respectively. Virions are remarkably stable and withstand extremes of ionic conditions, temperatures up to 55 °C, pH values between 2 and 9, lipid solvents, and detergents. Exposure to UV irradiation reduces infectivity.
Nucleic acid
All orthoreovirus genomes consist of 10 linear dsRNA segments that range from 0.60×106 to 2.60×106 Mr. The total Mr of the genome of the mammalian orthoreovirus Type 3 Dearing (Reovirus T3D or MRV-3De) isolate of Orthoreovirus mammalis is about 1.5×107 (23,549 bp) and constitutes approximately 11.5% of the virion mass. Based on their resolution by gel electrophoresis, the genomic dsRNAs are grouped into three length classes commonly referred to as long (L1–L3, about 3.9–3.8 kbp), medium (M1–M3, about 2.3–2.2 kbp), and short (S1–S4, about 1.6–0.9 kbp). The gel mobilities of certain genome segments are characteristic of some orthoreovirus species. For example, in comparison to MRV, the trigenic S1 genome segments of isolates of Orthoreovirus avis, Orthoreovirus neoavis and Orthoreovirus nelsonense (referred to colloquially as avian reoviruses [ARV], neoavian reoviruses [NARV] and Nelson Bay reoviruses [NBV], respectively) have reduced gel mobilities. Similarly, for isolates of Orthoreovirus papionis, Orthoreovirus mahlapitsiense and the “classical” Muscovy duck isolates of ARV (ARV-MdC) the equivalent segment is truncated and migrates as S4 (Duncan et al., 2004, Yun et al., 2014, Jansen van Vuren et al., 2016).
Complete virions contain numerous oligonucleotides (2–20 nt) representing approximately 25% of the total RNA content. Three-quarters of these are abortive reiterative 5′-terminal transcripts, produced by the reovirus core-associated transcriptase and capping enzymes, while the remainder are oligoadenylates. The 5′-terminus of the positive-sense RNA strand of each genome segment contains a dimethylated cap 1 structure (m7GpppGm2'OH). The genomic RNAs lack poly(A)-tails and do not contain covalently linked proteins. Genomic dsRNA segments contain 5′- and 3′-terminal sequences of 4 or 5 bp that are conserved in all 10 genome segments within a virus species. The 3′-terminal consensus sequence (UCAUC-3′) is conserved for members of all orthoreovirus species. In contrast, the 5′-terminal sequences are conserved within members of a species but vary between species and represent one of the polythetic criteria useful for assigning new isolates to a species (Table 1 Orthoreovirus).
Table 1 Orthoreovirus Conserved terminal sequences (positive-sense strand) of orthoreovirus genome segments
| Virus species | General name for viruses in the species | 5′-end | 3′-end |
|---|---|---|---|
| Orthoreovirus mammalis | mammalian reoviruses (MRV) | 5′-GCUA | UCAUC-3′ |
| Orthoreovirus avis | avian reoviruses (ARV) | 5′-GCUUUUU | UCAUC-3′ |
| Orthoreovirus nelsonense | Nelson Bay reoviruses (NBV) | 5′-GCUUUA | UCAUC-3′ |
| Orthoreovirus papionis | baboon reoviruses (BRV) | 5′-GUAAAUUU | UCAUC-3′ |
| Orthoreovirus reptilis | reptilian reoviruses (RRV) | 5′-GUUA/CUUUU | UCAUC-3′ |
| Orthoreovirus mahlapitsiense | Mahlapitsi reoviruses (MAHLV) | 5′-GGUCA | UCAUC-3′ |
| Orthoreovirus piscis | piscine orthoreoviruses (PRV) | 5′-GAUAA | UCAUC-3′ |
| Orthoreovirus broomense | Broome reovirus (BrRV) | 5-GUCAA | UCAUC-3′ |
| Orthoreovirus neoavis | neoavian reoviruses (NARV) | 5′-GCCUUUC | UCAUC-3′ |
| Orthoreovirus testudinis | testudine reoviruses (RRVT, ie reptilian reoviruses, testudine) | 5’-GUUA/C UUC | UCAUC-3′ |
Proteins
Orthoreovirus proteins are designated in terms of their relative length classes (λ, µ, and σ) and lengths (alphanumeric labeling) (Table 2 Orthoreovirus). The following discussion refers to the nomenclature scheme for mammalian orthoreovirus 3 Dearing strain T3D, the examplar isolate of the species Orthoreovirus mammalis.
Table 2 Orthoreovirus Genome segments and protein products of mammalian orthoreovirus 3 Dearing strain T3D
| Genome segment | bp | Protein | Mass (kDa) | Copies/ particle | Location and name | Function |
|---|---|---|---|---|---|---|
| L1 | 3854 | λ3 | 142 | 12 | Core, RdRP | RNA-directed RNA polymerase |
| L2 | 3916 | λ2 | 144 | 60 | Core turret | Capping enzyme with guanylyl and methyl transferase activity; turret protein |
| L3 | 3896 | λ1 | 143 | 120 | Core shell | Binds dsRNA; NTPase; helicase |
| M1 | 2304 | µ2 | 83 | 12 | Core NTPase | NTPase; inclusion body development; cell tropism; modulation of cellular interferon response |
| M2 | 2203 | µ1 µ1C | 76 72 | 600 | Outer shell | µ1 is cleaved to µ1C and myristolyated µ1N after assembly; during viral entry µ1C is further cleaved to δ and φ; µ1C multimerizes with σ3 in a T=13 symmetry in the outer capsid |
| M3 | 2235 | µNS µNSC | 80 75 | 0 | NS | Binds ssRNA and cytoskeleton; nucleates viral inclusion bodies; µNSC (unknown function) derived from alternate translation start site |
| S1 | 1416 | σ1 | 49 | 36 | Outer fiber | Viral attachment protein; homotrimer; cell tropism; pathways of viral spread in vivo |
| σ1s | 16 | O | NS | Viral spread in vivo; cell cycle arrest | ||
| S2 | 1331 | σ2 | 47 | 150 | Core clamp | Inner capsid structural protein assumes a T=1 symmetry |
| S3 | 1189 | σNS | 41 | 0 | NS | RNA stability; ssRNA-binding; inclusion body development |
| S4 | 1196 | σ3 | 41 | 600 | Outer clamp | dsRNA-binding, multimerizes with µ1; nuclear and cytoplasmic localization; translation control; modulation of cellular interferon response |
Virions contain eight different structural proteins. The stabilizing lattice of the outer capsid is composed of 200 interlocking trimers of the 76 kDa µ1 outer shell protein. The µ1 subunits also interact with monomers of the σ3 outer-capsid clamp protein, which represent finger-like projections on the surface of the virion (Liemann et al., 2002). Pentameric subunits of the λ2 protein make up the flower-like structures and turrets at the vertices of viral particles and cores, respectively. The λ2 structures interact with subunits of the tetrameric σ3 clusters and with the µ1 lattice and represent essential structural components of the outer capsid. This outer-capsid protein remains associated with core particles, unlike the other outer-capsid proteins. The fourth component of the outer capsid, the σ1 fiber protein, exists as 12 homotrimers associated with the vertices of virions and ISVPs (Chappell et al., 2002). It may assume either a retracted or extended conformation. The λ1 (120 copies) core shell and σ2 core clamp (150 copies) proteins represent the major structural proteins of the inner capsid. The final two structural proteins of the virus, the λ3 RdRP and the µ2 NTPase, are present at about 12 copies per virion and are located on the inside of the inner capsid. The λ3 protein forms 7 nm projections that extend toward the interior of the core, underlying the 12 vertices of the capsid. The µ2 protein may be associated with these λ3 structures. The genome also encodes 3-6 nonstructural proteins (NS) (Table 2 Orthoreovirus), including the FAST proteins responsible for syncytium formation (Figure 2 Orthoreovirus).
Lipids
Mature virions lack a lipid envelope. The major outer capsid lattice protein, µ1, and its µ1N cleavage product are N-terminally myristoylated (Nibert et al., 1991). The FAST protein responsible for syncytium formation induced by the fusogenic reoviruses are either N-terminally myristoylated or palmitoylated at internal cysteine residues (Ciechonska and Duncan 2014). These acylations are essential for the membrane fusion activity of the proteins.
Carbohydrates
Convincing evidence that any of the orthoreovirus proteins are glycosylated has not been reported. Moreover, no carbohydrate has been observed in the structures of any of the mammalian reovirus proteins that have been determined by X-ray crystallography (λ1, λ2, λ3, µ1, σ1, σ2, and σ3).
Genome organization and replication
The genome consists of ten segments of linear dsRNA, which are packaged in equimolar ratios (one copy of each within each virion). The segments possess terminal non-translated regions (NTRs) that are shorter at the 5′-terminus (12–32 bp for MRV3) than at the 3′-terminus (35–85 bp). The major ORFs vary in length from 1059 to 3867 bp. All orthoreoviruses contain one S-class genome segment that encodes more than one ORF and that has undergone considerable genetic rearrangement among viruses within and between species. The MRV S1 segment is bigenic, encoding the σ1 protein and the σ1s protein from a second embedded ORF (Table 2 Orthoreovirus and Figure 2 Orthoreovirus). Isolates of Orthoreovirus piscis, Orthoreovirus reptilis and Orthoreovirus testudinis (collectively referred to as piscine reoviruses [PRV], reptilian reoviruses [RRV] and testudines reptilian reoviruses [RRVT], respectively) also have bigenic S1 genome segments but with different gene arrangements. PRV encodes the σB outer-clamp protein and a cytotoxic p13 protein that is presumably nonstructural and has no sequence similarity with MRV σ1s or with any of the other small orthoreovirus NS proteins (Key et al., 2013). A different arrangement is observed for the S1 genome segment of isolates of Orthoreovirus reptilis and Orthoreovirus testudinis in which there are two sequential, partially overlapping ORFs encoding a σC fiber protein homolog and a p14 FAST protein. The S1 genome segments of ARV and NBV are trigenic, encoding the fiber protein σC, the p17 protein implicated in cell cycle arrest and autophagy induction (Huang et al., 2017) and the homologous p10 FAST proteins responsible for virus-induced syncytium formation. The S1 genome segment of “novel” Muscovy duck ARV isolates (ARV-MdN) shares this trigenic arrangement, although the p10.2 protein does not function as a FAST protein (Wang et al., 2012, Yang et al., 2020). This arrangement differs from that of “classical” Muscovy duck reoviruses (ARV-MdC) that contain a truncated S1 genome segment-equivalent (S4, 1124 bp) encoding a σC fiber protein homolog and a p10.8 protein that is unrelated to the p10 FAST proteins and which localizes to the nucleus and induces apoptosis (Geng et al., 2009, Guo et al., 2014). BRV and MAHLV lack a σ1/σC fiber homolog and contain bigenic S4 genome segments encoding homologous p16 NS proteins of unknown function and unique p14 and p15 FAST proteins (Figure 2 Orthoreovirus).
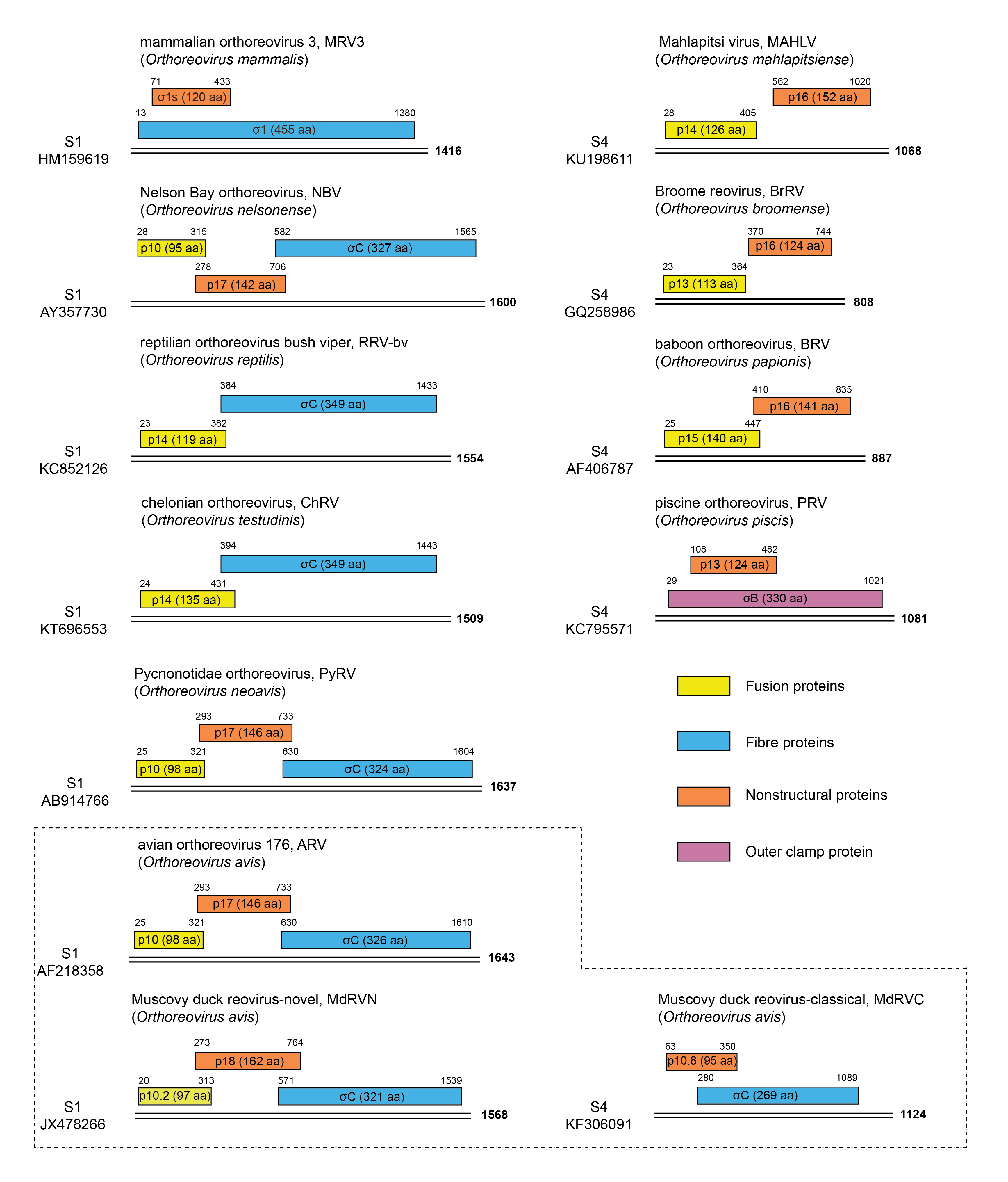 |
| Figure 2 Orthoreovirus Gene organization of the polycistronic genome segments of members of the ten species of orthoreoviruses. Double lines represent the genomic dsRNAs, with segment lengths indicated at the right hand ends. ORFs are shown as colour coded rectangles with numbers indicating their first and last nucleotides (including the termination codons). The identities and lengths in amino acid residues of the gene products encoded by the various ORFs are indicated within the rectangles. Virus names, abbreviations and virus species are indicated, along with genome segment (S1 or S4) and the corresponding GenBank accession. The dotted lines separate three isolates of the species Orthoreovirus avis; those isolated from Muscovy ducks, referred to as the “novel” (MdRVN) have a trigenic gene arrangement while “classical” (MdRVC) isolates have a bigenic gene arrangement, and their S1 segment migrates as S4. |
The overall course of infection involves virion adsorption, low pH-dependent penetration and uncoating to core particles, asymmetric transcription of capped, non-polyadenylated mRNAs via a fully conservative mechanism (the nascent strand is displaced), translation, assembly of positive-sense strands into progeny subviral particles, conversion of positive-sense strands to dsRNA, and further rounds of mRNA transcription and translation. The efficiency of translation of the various reovirus mRNAs varies over a 100-fold range, whereas the proportions of the mRNAs found in infected cells vary inversely to their length. The final stage of the replication cycle involves the assembly of the outer capsid onto progeny subviral particles to form infectious virions. Based on studies of MRV replication, virion morphogenesis is thought to proceed along a pathway involving a series of assembly intermediates. Progeny particles accumulate in paracrystalline arrays in the perinuclear region of the cytoplasm and are released from some cells following lysis late in the replication cycle. Reovirus egress from other cells is nonytic (Lai et al., 2013). In addition to the generalized replication cycle, the fusogenic orthoreoviruses induce formation of multinucleated syncytia that enhance direct cell-cell transmission of the infection and the kinetics of virion release, thereby contributing to the virulence of the fusogenic reoviruses following natural infections (Duncan and Sullivan 1998, Salsman et al., 2005).
The functions and properties of specific viral proteins influence various stages of the MRV replication cycle (Table 2 Orthoreovirus). The reovirus σ1 fiber attachment protein determines the cell and tissue tropism of the virus strain and has hemagglutination activity. The σ1 protein binds cell-surface carbohydrate and junctional adhesion molecule-A (Barton et al., 2001). An additional receptor, NogoR1, has been implicated in MRV3 infection (Konopka-Anstadt et al., 2014). However, the viral protein that engages NogoR1 has not been determined. The µ1 outer-capsid protein is N-terminally myristoylated and forms a complex with the σ3 outer clamp protein in solution that triggers cleavage of µ1 to µ1N and µ1C (Odegard et al., 2004). The µ1C fragment is further proteolytically cleaved into δ and φ polypeptides during virion entry into cells and is responsible for membrane penetration (Nibert and Fields 1992). The µ1 outer-capsid protein also influences strain-specific differences in capsid stability, transcriptase activation, apoptosis, and neurovirulence. In the case of ARV, the µ1 homolog (µB) is implicated in strain-specific differences in macrophage infection (O'Hara et al., 2001). In addition to interacting with µ1 and forming the outer capsid layer of the virion, the σ3 outer clamp protein is a dsRNA-binding protein involved in translation regulation, altering the activity of protein kinase R, and modulating the interferon response (Sherry 2009). The λ2 core turret is the guanylyltransferase involved in mRNA capping, while the λ1 core shell and σ2 core clamp proteins both bind dsRNA. The λ1 core protein also may function as a helicase and an RNA triphosphatase. The λ3 RdRP and the µ2 NTPase are minor core proteins.
All orthoreoviruses also encode two major NS proteins (Table 2 Orthoreovirus). The µNS and σNS proteins are produced in high abundance during infection and, together with σ3, associate with mRNA to form virus mRNA-containing complexes, which are presumed to be precursors of progeny virion assembly. The σNS protein binds and stablilizes ssRNA, disrupts stress granules, and along with µNS generates intracytoplasmic inclusions called “virus factories” that are the sites of virion macromolecular synthesis and assembly (Choudhury et al., 2017). The µNS protein also associates with the core NTPase µ2 that stabilizes microtubules within viral inclusions (Carvalho et al., 2007). Additional NS proteins encoded on the polygenic genome segments of isolates of different Orthoreovirus species (Figure 2 Orthoreovirus) include the FAST proteins, σ1s involved in virion dissemination in infected mice and cell-cycle arrest (Boehme et al., 2009), the p17 proteins that induce cell cycle arrest and autophagy, and several other small NS proteins of undefined function (Figure 2 Orthoreovirus).
Replication strategies used by the fusogenic reoviruses such as ARV, NBV, RRV, MAHLV and BRV are similar to that described above for the prototypical MRV, with some notable exceptions. In addition to the generalized replication cycle, the fusogenic reoviruses encode a FAST protein that induces formation of multinucleated syncytia to enhance direct cell-cell transmission of the infection and the kinetics of virion release, thereby contributing to the virulence of the fusogenic orthoreoviruses following natural infection. The homologous p10 FAST proteins of ARV, NARV and NBV share sequence and structural similarities. The p14 FAST proteins of RRV, RRVT and MAHLV and the p13 FAST protein of BrRV share detectible, but limited, sequence conservation. The p10 and p14 FAST proteins appear unrelated to each other and to the BRV p15 FAST protein. All these FAST proteins are small, basic, acylated, bitopic transmembrane proteins (type I membrane topology) and induce fusion in transfected cells in the absence of other viral proteins (Ciechonska and Duncan 2014). The truncated fiber proteins of the fusogenic orthoreoviruses contain a trimeric coiled-coil domain and globular head like that of MRV σ1 but are shorter due to species-specific internal indels in the coiled-coil and β-spiral repeat regions (Yang et al., 2020). BRV, BrRV, PRV, and MAHLV encode no homolog of the fiber viral attachment protein and presumably use features in their surface exposed capsid proteins to mediate cell attachment. The dsRNA-binding domain of the MRV σ3 outer-capsid protein is not conserved in the homologous σB proteins of ARV, NBV, or BRV. However, as with the MRV-3 σ2 core protein, the ARV σA core protein displays dsRNA-binding activity and may function analogously to the MRV3 σ3 outer-capsid protein to regulate protein kinase R activity and the interferon response (Lostalé-Seijo et al., 2016).
Biology
Transmission of orthoreoviruses is by an enteric or respiratory route; no arthropod vectors are involved, and infection is restricted to a wide variety of vertebrate species (baboons, bats, birds, cattle, humans, monkeys, sheep, snakes, swine, and rodents). Orthoreovirus distribution is worldwide. Human orthoreoviruses generally do not produce symptoms but may cause upper respiratory tract illness and possibly, rarely, enteritis in infants and children. Strains of certain orthoreoviruses can abrogate immunologic tolerance to newly introduced food antigens and may induce celiac disease in humans (Bouziat et al., 2017). In laboratory mice, orthoreovirus infection can cause diarrhea, runting, oily hair syndrome, hepatitis, jaundice, myocarditis, myositis, pneumonitis, encephalitis, and hydrocephalus. A variety of signs may be associated with orthoreovirus infection of domestic animals including upper and lower respiratory tract illnesses and diarrhea. In rhesus monkeys, orthoreoviruses cause hepatitis, extrahepatic biliary atresia, meningitis, and necrosis of ependymal and choroid plexus epithelial cells. The BRV isolate (BRV-USA/11-1993) was obtained from yellow baboons with meningoencephalomyelitis (Duncan et al., 1995, Kumar et al., 2014). RRV-bv isolates from a green bush viper and a python were obtained from animals displaying respiratory or neurological signs (Lamirande et al., 1999, Vieler et al., 1994). The outcome of ARV infection in birds ranges from inapparent to lethal, depending on the virus strain and the age of the host. Systemic infection results in virus dissemination to numerous tissues. Disease presentations in chickens include feathering abnormalities, gastroenteritis, hepatitis, malabsorption, myocarditis, paling, pneumonia, stunted growth and weight loss (Jones 2000). In turkeys, ARV causes enteritis. Birds that survive an acute systemic infection may develop joint and tendon disorders (tenosynovitis) that resemble the pathology of rheumatoid arthritis in humans. PRV is associated with heart and skeletal muscle inflammation in farmed salmon and contributes to substantial economic losses in the aquaculture industry (Palacios et al., 2010).
MRV and ARV induce the biochemical and morphologic hallmarks of apoptosis in cultured cells (Danthi et al., 2013). MRV infection leads to activation of nuclear factor kappa B (NF-κB) (Connolly et al., 2000), a family of transcription factors that play important roles in regulating cellular stress responses, including apoptosis. The µ1 cleavage fragment φ, which is released following disassembly, is an important trigger of NF-κB activation (Danthi et al., 2008), but the precise mechanism is unclear. Apoptosis induced by orthoreovirus infection requires both extrinsic (death-receptor) and intrinsic (mitochondrial) signaling pathways linked by the small Bcl-2 family member, Bid (Danthi et al., 2010). Another Bcl-2 family member, Noxa, is also implicated (Knowlton et al., 2012). As with MRV3, ARV-induced apoptosis requires virion disassembly but not viral transcription. In some cultured cell types, MRV3 induces cell death by regulated necrosis (necroptosis) (Berger and Danthi 2013). Necroptosis induction requires IFN signaling and RIP kinase 3 and depends on viral transcription.
MRV3 preferentially replicates in a lytic manner in transformed cells, due to the combined inhibitory effects of cell transformation on innate immune signaling pathways and enhancing effects on cell metabolism, providing the basis for development of MRV3 as an oncolytic agent for cancer therapy (Gujar et al., 2018).
Antigenicity
The serotype-specific antigen of MRV and ARV is the fiber protein, which is recognized by neutralizing antibodies (Weiner and Fields 1977). Antigenic recognition of this protein was the basis for the original serotyping of MRV and ARV isolates. The MRV σ1 and σ1s proteins elicit strain-specific and cross-reactive cytotoxic T-cell activities. The MRV proteins λ2 and σ3 are species-specific antigens, like the λB and σB proteins of ARV. The considerable sequence similarity that exists between different isolates of the same orthoreovirus species, but not between species, is reflected by the limited antigenic cross-reactivity detected between members of different species. The most extensive antigenic similarity between members of different species occurs between ARV and NBV, in accordance with the significant amino acid sequence identity between viruses of these species.
Species demarcation criteria
There are ten species of orthoreoviruses. The classification of viruses into these species is supported by experiments showing reassortment of genome segments between isolates of the same species but not between those of different species. In addition to the other general criteria used throughout the family, members of a species in the genus Orthoreovirus may be identified by:
- Extensive sequence identity between the proteins encoded by homologous genome segments (for most conserved core proteins, >85% amino acid sequence identity within a species versus <65% identity between species; for most of the more divergent outer capsid proteins, >55% identity within a species and <35% between species). Viruses of the ten species of orthoreovirus represent evolutionarily distinct lineages, as illustrated by phylogenetic analysis using the amino acid sequences of the outer-clamp protein, for which the greatest number of sequences from diverse isolates is available (Figure 3 Orthoreovirus).
- Extensive sequence identity between homologous genome segments (for most genome segments, >75% nucleotide sequence identity within a species versus <60% between species)
- Conserved 5′-terminal sequences on most if not all genome segments within a species
- Similar organization of the multigenic genome segment
- Analysis of electropherotype by agarose gel electrophoresis but not by polyacrylamide gel electrophoresis (some similarities can exist between closely related species)
- Identification of host species and disease signs
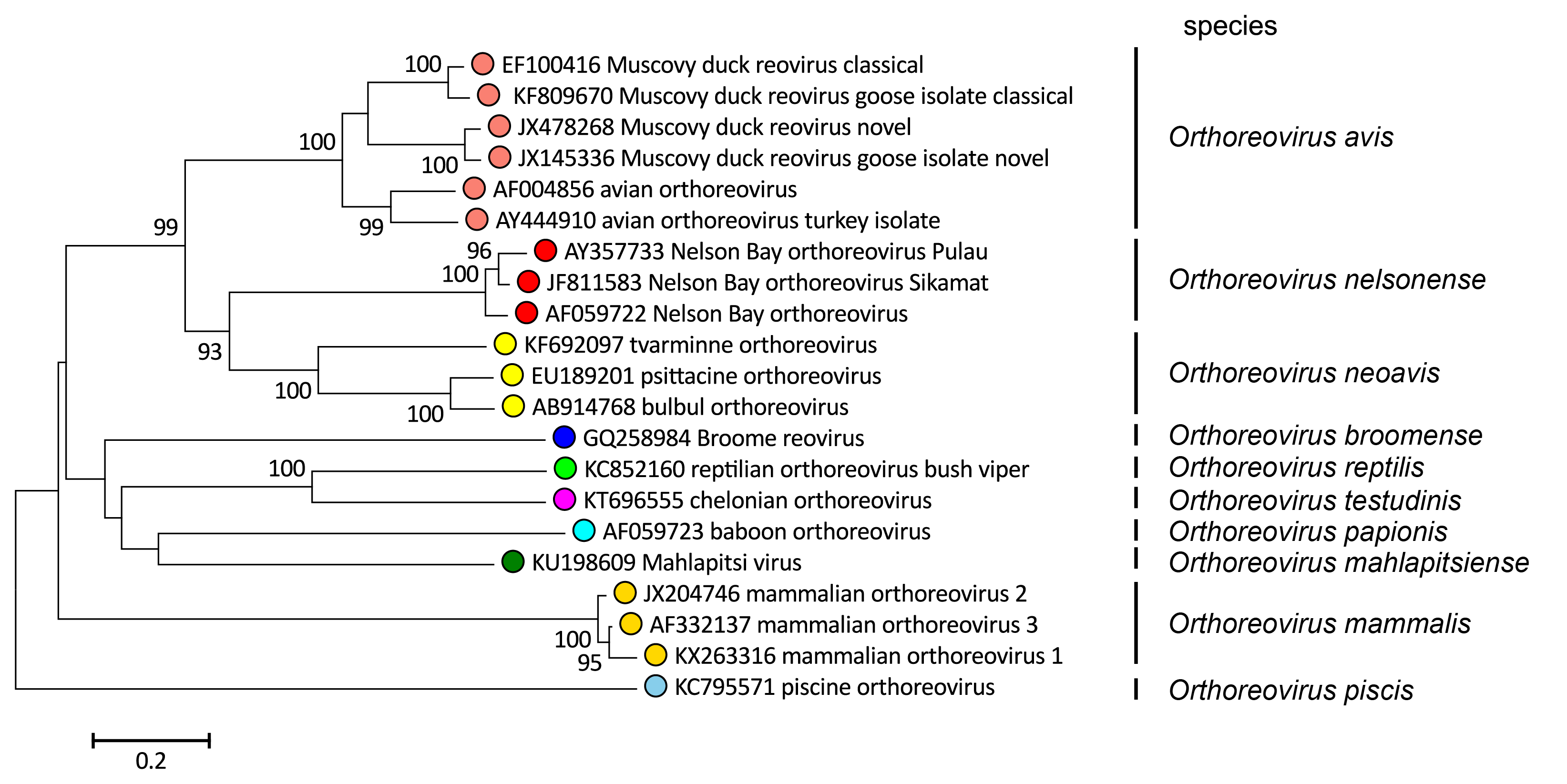 |
| Figure 3 Orthoreovirus Phylogenetic relationships of orthoreovirus outer clamp proteins. Sequences were aligned using MUSCLE (Edgar 2004) and the unrooted neighbour-joining tree constructed using MEGA 7 (Kumar et al., 2016). The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) where this was >70 % are shown next to the branches. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Orthoreovirus mammalis includes viruses which can be distinguished as three major serotypes (MRV1, MRV2, and MRV3) each comprising numerous isolates, and a fourth serotype (MRV4) with only one isolate, Ndelle reovirus. Amino acid sequence identities of the sigma-class major outer-capsid and core proteins of various serotypes range from 90 to 97%.
Orthoreovirus avis includes numerous isolates from commercial landfowl and waterfowl, including chickens, Muscovy ducks, turkeys, and geese. Sequence diversity is more extensive among the various ARV isolates than among MRV isolates (68–95% in the outer-clamp protein). There also is considerable divergence in the polygenic genome segments between some Muscovy duck and goose isolates and the prototypical ARV isolates (Figure 2 Orthoreovirus).
Orthoreovirus neoavis includes isolates obtained from a hooded crow (Corvus corone cornix) (Huhtamo et al., 2007), different species of psittacine birds (de Kloet 2008), and a brown eared bulbul (Hypsipetes amaurotis) (Ogasawara et al., 2015). Collectively, these isolates are referred to as neoavian reoviruses (NARV). Infected animals show diverse neurological, respiratory and mutli-organ pathology. These isolates share a 5′-terminal heptanucleotide sequence (5′-GCUUUUC) that differs in the seventh position (5′-GCUUUUU) from the waterfowl and landfowl isolates of Orthoreovirus avis (Table 1 Orthoreovirus). Amino acid identities in their outer-clamp protein are 55−56% versus 37−39% when compared to the homologous protein of isolates of the species Orthoreovirus avis.
Orthoreovirus nelsonens includes several syncytium-inducing mammalian reovirus isolates from fruit bats, some of which were isolated from humans with severe respiratory infections suggesting zoonotic transmission of these viruses. The sequence similarity between NBV and ARV exceeds that between NBV and members of other orthoreovirus species. NBV and ARV display a similar gene organization of the polygenic S1 genome segment, and they encode homologous p10 FAST proteins (Figure 2 Orthoreovirus). Although ARV and NBV clearly share a more recent evolutionary past than the viruses of other orthoreovirus species, the extent of sequence divergence (59–61% identity in the core-clamp protein and only 29–36% identity in the outer clamp protein) and the unique 5′-terminal genome segment sequences (Table 1 Orthoreovirius) justify the designation of NBV as a member of a separate species.
Orthoreovirus papionis includes a single sequenced isolate, BRV, obtained from the brains of yellow baboons diagnosed with meningoencephalomyelitis. BRV shares little sequence (16–32% amino acid sequence identity between homologous S-class gene products) or antigenic similarity with the other fusogenic species, it contains a polygenic S4 genome segment with a distinct gene organization, a unique p15 FAST protein, and a unique 5′-terminal consensus sequence, all of which identify BRV as belonging to a distinct species.
Orthoreovirus testudinis includes a single isolate obtained from a spur-thighed tortoise (Testudo graeca Linnaeus, 1758) in Switzerland with pathologic indicators of cachexy, tongue epithelial necrosis and splenomegaly (Kugler et al., 2016). Genome segments contain a conserved 5′-terminal sequence (5′-GUUA/CUUC) distinct from that of members of the species Orthoreovirus reptilis (Table 1 Orthoreovirus). The outer-clamp protein shares 45% amino acid identity with the homologous protein from of members of the species Orthoreovirus reptilis. Together, these criteria identify Orthoreovirus testudinis as a representative of a distinct species of reptilian orthoreoviruses.
Orthoreovirus reptilis includes isolates from snakes and lizards. Complete genomic sequence information is available for an isolate from a green bush viper (RRV-bv), and partial sequences are available for a python isolate (RRV-py) and several isolates from lizards. Amino acid sequence identities between the RRV outer-clamp protein and the homologous protein of viruses of other species are 16–25%. RRV possesses a unique 5′-terminal sequence (Table 1 Orthoreovirus) and a unique gene arrangement on the bigenic S1 genome segment (Figure 2 Orthoreovirus). It encodes a p14 FAST protein that shares some sequence identity (31%) with the p14 FAST protein of MAHLV but not with other FAST proteins.
Orthoreovirus piscis includes numerous closely related isolates from salmonid fish. PRV has only 13–18% amino acid identity with the outer-clamp protein of members of other species, and PRV genome segments contain a unique 5′-terminal consensus sequence. PRV also has a unique bigenic S1 genome segment encoding the outer-clamp protein and a small NS cytotoxic protein. It does not encode a FAST protein and is hence the only nonfusogenic orthoreovirus apart from MRV. Phylogenetic trees based on concatenated sequences of the nine homologous proteins (seven structural and two nonstructural proteins) show PRV clustering more closely with orthoreoviruses than with aquareoviruses. In addition, PRV contains 10 genome segments, not 11 as with aquareoviruses (Nibert and Duncan 2013). Based on this evidence, PRV was assigned as member of a new species of orthoreovirus, and the only species whose members are currently known to infect fish.
Mahlapitsi virus (MAHLV), the sole isolate of the species Orthoreovirus mahlapitsiense was isolated from a fly (Eucampsipoda africana Theodor, 1955) associated with fruit bats and is the only orthoreovirus isolated from an arthropod, although it is possible that this may have been a passive host for the virus. MAHLV has a bigenic S4 genome segment encoding homologues of the RRV p14 FAST protein and BRV p16 NS protein and genome segments contain a unique 5′-terminal consensus sequence (Table 1 Orthoreovirus). Amino acid sequence identities between the MAHLV outer-clamp protein and the homologous protein of other orthoreoviruses are only 6–24%.
Orthoreovirus broomenseincludes a single isolate obtained from a little red flying fox (Pteropus scapulatus Peters, 1862) with neurological symptoms, found in Broome, Australia (Thalmann et al., 2010). The BrRV outer-clamp protein shares <27% amino acid identity with the homologous protein from other orthoreovirus species and has a conserved 5′-sequence that is distinct from the conserved terminal sequences in other species. As with PRV, BRV and MAHLV, BrRV lacks a fiber protein and contains a bigenic S4 genome segment encoding a homologous p16 protein of no defined function and a p13 FAST protein most closely related to the p14 FAST proteins of RRV-bv and MAHLV.
Related, unclassified viruses
| Virus name | Accession number | Abbreviation |
| Steller sea lion reovirus | L1: HM222978; L2: HM222980; L3: HM222979; M1: HM222977; M2: HM222976; M3: HM222975; S1: HM222974; S2: HM222973; S3: HM222972; S4: HM222971 | SSRV |
Virus names and virus abbreviations are not official ICTV designations.

