Family: Sedoreoviridae
Jelle Matthijnssens, Houssam Attoui, Krisztián Bányai, Corina P. D. Brussaard, Pranav Danthi, Mariana del Vas, Terence S. Dermody, Roy Duncan, Qín Fāng (方勤), Reimar Johne, Peter P. C. Mertens, Fauziah Mohd Jaafar, John Patton, Takahide Sasaya (笹谷孝英), Nobuhiro Suzuki (鈴木信弘) and Taiyun Wei (魏太云)
The citation for this ICTV Report chapter is the summary published as Matthijnssens et al., (2022) ICTV Virus Taxonomy Profile: Sedoreoviridae, Journal of General Virology (2022) 103:001782.
Corresponding author: Jelle Matthijnssens (jelle.matthijnssens@kuleuven.be)
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: October 2021, June 2022
Summary
The family Sedoreoviridae includes viruses that have relatively smooth core particles, in contrast to members of the family Spinareoviridae which have large spikes or turrets situated at the 12 icosahedral vertices of either the virion or core particle (Table 1.Sedoreoviridae). The transcriptionally active core particle of sedoreovirids is made of two protein layers. The inner layer contains 120 copies of a single protein with a T=2 symmetry. The outer contains 780 copies of another single protein with a T=13 symmetry. The outer capsid (usually made of 2–3 proteins species) surrounding the core particle, shows greatest diversity in both the sequences of its components and/or structural organisations. However, sequence and structural analyses depicted similar structural organisations for outer capsid protein of rotaviruses and seadornaviruses (Mohd Jaafar et al., 2005).
Table 1.Sedoreoviridae Characteristics of members of the family Sedoreoviridae
| Characteristic | Description |
| Example | Rotavirus A RVA/Simian-tc/ZAF/SA11-H96/1958/G3P5B[2] (Seg1: DQ838640; Seg2: DQ838635; Seg3: DQ838645; Seg4: DQ841262; Seg5: DQ838599; Seg6: DQ838650; Seg7: DQ838610; Seg8: DQ838615; Seg9: DQ838620; Seg10: DQ838625; Seg11: DQ838630), species Rotavirus alphagastroenteritidis |
| Virion | Non-enveloped, icosahedral, 60–100 nm virions composed of 1–3 concentric capsid proteins layers. |
| Genome | 18–26 kbp of segmented linear dsRNA, with each of the 10–12 segments ranging from 0.6–5.8 kbp. |
| Replication | Replication occurs in the cytoplasm in electron-dense structures called viroplasms or virus inclusion bodies. |
| Translation | From full-length transcribed mRNAs, which possess a 5′-terminal cap but no poly(A)-tail. |
| Host Range | Mammals, birds, crustaceans, arthropods, algae and plants (Mohd Jaafar et al. 2014, Suzuki et al. 2015). |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Duplornaviricota, class Resentoviricetes, order Reovirales: 7 genera and 45 species |
Virion
Sedoreovirid particles are icosahedral. The protein capsid is organized as 1–3 concentric layers of capsid proteins, with an overall diameter of 60–100 nm (Mohd Jaafar et al., 2005). Members of Sedoreoviridae lack large surface projections on the subviral particle, giving them an almost spherical or “smooth” appearance (Fig. 1), in contrast to members of the Spinareoviridae, which have spikes or turrets at the 12 icosahedral vertices.
Genome organisation and replication
Sedoreovirids contain 10–12 segments of linear dsRNA comprising 18–26 kbp in total, with individual segments ranging from 0.6–5.8 kbp. The positive-sense strands of each duplex are modified with a 5′-terminal type 1 cap structure but no 3′-poly(A) tail. The viral RNAs are mostly monogenic with relatively short 5′- and 3′-non-coding regions, although some segments have a second or third functional open reading frames (Belhouchet et al., 2011).
Virus cell entry varies between genera but usually results in loss of outer-capsid components. The resulting transcriptionally-active particles are released into the cytoplasm. The 5′-capped mRNAs are synthesized by structural enzymatic components of the particle and are released through pores at the icosahedral apices of the virion into the cytoplasm. Viroplasms, also known as virus inclusion bodies, are distributed throughout the cytoplasm. These neo-organelles are sites of viral mRNA synthesis, genome replication, and particle assembly (Papa et al., 2021). Sets of a single copy of each capped mRNA are incorporated into progeny virus particles (Boyce et al., 2016). These mRNAs serve as templates for negative-sense strand synthesis, thereby reconstituting genomic encapsidated dsRNAs. The steps involved in virion morphogenesis and virus egress from cells differ depending on the genus (Trask et al., 2012). Progeny virions are released from some cell types without compromising cell viability (e.g., budding), or from other types of cells following cell lysis (Mohd Jaafar et al., 2005).
Biology
Host are mammals, birds, crustaceans, arthropods, algae and plants (Mohd Jaafar et al., 2014, Suzuki et al., 2015). The biological properties of the viruses vary according to genus. See individual genus sections for details.
Derivation of names
Cardoreovirus: from Carcinus, latinized from the Ancient Greek Καρκινος meaning crab, and
Mimoreovirus: from Micromonas, the genus name of the protist host from which virus was first isolated.
Orbivirus: from orbis, Latin for “ring” or “circle”, in recognition of the ring-like structures observed in micrographs of the surface of core particles.
Phytoreovirus: from phyton, Greek for “plant”.
Rotavirus: from rota, Latin for “wheel”.
Seadornavirus: from South-East Asian dodeca (from the Ancient Greek δωδεκα, meaning twelve) RNA viruses.
Sedoreoviridae: from sedo, Latin for “smooth”, denoting a smooth appearance and the absence of spikes or turrets on the surface of the core particles.
Genus demarcation criteria
The number of genome segments (10, 11, or 12) is in most cases characteristic of viruses within a single genus.Host (and vector) range and disease signs also are important indicators that help to identify viruses from different genera. Capsid structure (number of capsid layers, and the symmetry and structure of the outer capsid) also can be significant. The level of sequence divergence, particularly in the more conserved genome segments and proteins (for example as detected by comparisons of RdRP or inner-capsid-shell proteins and the segments from which they are translated) can be used to distinguish members of different genera (Attoui et al., 2006). Available data suggest that isolates from different genera usually have <26% amino acid identity in comparisons between their RdRPs, while within a single genus, identities are usually >30% (Attoui et al., 2002).
Species demarcation criteria
The prime determinant for inclusion of virus isolates within a single virus species is their capacity to exchange genetic information during co-infection by genome segment reassortment, thereby generating viable progeny virus strains. However, data providing direct evidence of segment reassortment between isolates are available only for viruses of a few genera. The following methods are therefore commonly used (preferably in combination) to examine levels of similarity between isolates and to predict their possible compatibility as reassortants:
- Nucleotide and amino acid sequence analysis (viruses within different species should have low levels of sequence similarity between cognate genome segments).
- Serological comparisons of antigens or antibodies using either polyclonal antisera or monoclonal antibodies against conserved antigens. Methods used may include ELISA, complement fixation, and agar gel immunodiffusion. Closely related isolates and serotypes generally belong to the same species.
- Analysis of electropherotype by agarose gel electrophoresis (AGE) but not by polyacrylamide gel electrophoresis (PAGE). Virus isolates within the same species will show a relatively uniform electropherotype. However, a major deletion/insertion event may sometimes result in two distinct electropherotypes within a single species, and apparent similarities can exist between more closely related species.
- Identification of the conserved terminal regions of the genome segments. These are usually conserved across all segments within a species, although some closely related species also can have identical terminal sequences of at least some segments.
These criteria apply throughout the family. Additional or more specific criteria are provided in the section for each genus, where applicable.
Relationships within the family
Members of different genera segregate into different clades upon phylogenetic analysis of the RdRP protein (Figure 1.Sedoreoviridae)
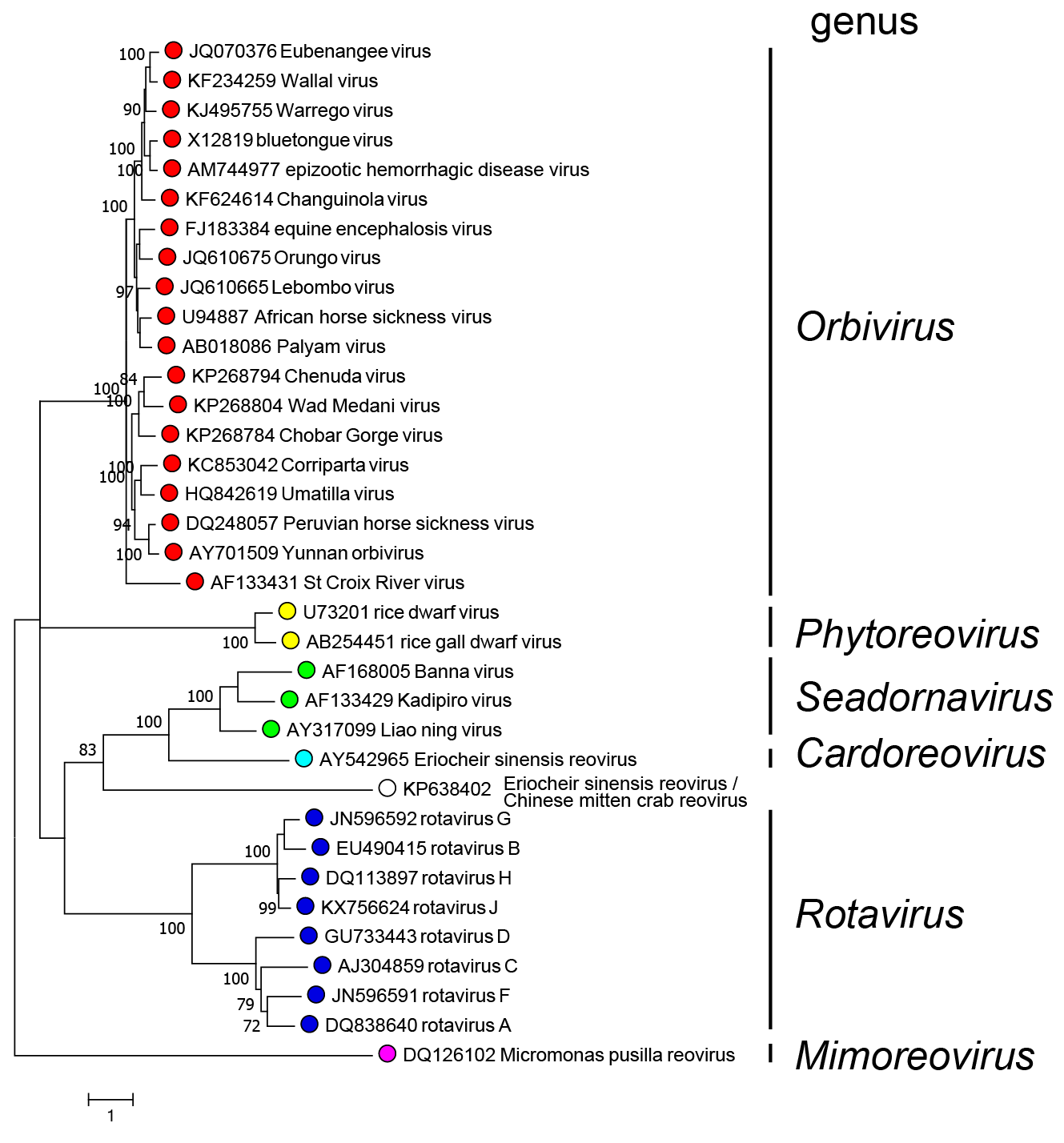 |
| Figure 1.Sedoreoviridae. Phylogenetic tree of RdRP proteins of members of the family Sedoreoviridae. Maximum likelihood tree constructed in MEGA7 (Kumar et al., 2016) using the amino acid sequence of the putative RdRP of representative viruses in the family Sedoreoviridae. The analysis used the Le Gascuel 2008 + Freq. model with a discrete gamma distribution of rates between sites, including some invariant sites. Values at the nodes represent bootstrap confidence levels (100 replications). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Sedoreovirids are most closely related to the members of the family Spinareoviridae, both families being placed in the order Reovirales. For a discussion of relationships with more distantly related viruses see the discussion in the Reovirales Report.
Related, unclassified viruses
| Virus name | Accesssion number | Abbreviation |
| Eriocheir sinensis reovirus; Chinese mitten crab reovirus | Seg1: KP638402; Seg2: KP638403; Seg3: KP638404; Seg4: KP638405; Seg5: KP638406; Seg6: KP638407; Seg7: KP638408; Seg8: KP638409; Seg9: KP638410; Seg10: KP638411; Seg11: KP638412; Seg12: KP638413 | ESRV; CMCRV |
Virus names and virus abbreviations are not official ICTV designations.
The Eriocheir sinensis reovirus (ESRV) genome consists of 12 segments of linear dsRNA that are numbered in order of reducing Mr, or increasing electrophoretic mobility during agarose gel electrophoresis (AGE) (Table 2.Sedoreoviridae) and comprise approximately 23,300 bp in total. The AGE electropherotype follows a 3-4-2-3 pattern. The terminal sequences of ESRV Seg1 are 5′-GGAUUUAAAA ….AUAACAGAC-3′.
Table 2.Sedoreoviridae. Genome segments and protein products of ESRV
| Genome segment | bp | Protein | Protein mass (kDa)* | Structure/function |
|---|---|---|---|---|
| Seg1 | 4243* | VP1(Pol) | 138 | RdRP (transcription complex) |
| Seg2 | 2711 | VP2 | 100 | unknown |
| Seg3 | 2710 | VP3 | 97 | unknown |
| Seg4 | 2441 | VP4 | 50 | unknown |
| Seg6 | 2141 | VP6 | 67 | unknown |
| Seg5 | 2082 | VP5 | 61 | unknown |
| Seg7 | 1522 | VP7 | 46 | unknown |
| Seg10 | 1265 | VP10 | 37 | unknown |
| Seg8 | 1233 | VP8 | 33 | unknown |
| Seg11 | 1200 | VP11 | 23 | unknown |
| Seg12 | 1186 | VP12 | 30 | unknown |
| Seg9 | 1179 | VP9 | 38 | unknown |
* partial coding sequence; all other genome segment sequences are partial with a complete coding sequence.

