Family: Sedoreoviridae
Genus: Phytoreovirus
Distinguishing features
Phytoreovirus particles have icosahedral symmetry with a distinctive angular appearance and contain 12 dsRNA species. They are transmitted by cicadellid leafhoppers to susceptible plant species, replicating in both hosts and vectors.
The significant similarities among corresponding proteins of the phytoreoviruses and the absence of similarity to any other reoviruses proteins, confirm that viruses of the genus Phytoreovirus in the order Reovirales constitute a distinct group.
Virion
Morphology
Virions of rice dwarf virus (RDV) are icosahedral, appear to be double-layered and are about 70 nm in diameter (Figure 1.Phytoreovirus). The outer layer of RDV contains 260 trimers of P8 (46 kDa): a total of 780 molecules, arranged with T=13 symmetry (Figure 1.Phytoreovirus). The relative location of the neighboring capsomers on the icosahedral particle is such that they form pentameric or hexameric rings. The inner capsid layer is reported to be a complete protein shell, composed of 60 dimers of P3 (114 kDa), a total of 120 molecules, arranged with a suggested T=1 icosahedral symmetry (Figure 1.Phytoreovirus). The outer capsid P8 trimers bind more tightly at the threefold positions of the single-layered core. The RDV virion structure appears to be comparable to that of core or double-layered particles of viruses of the genera Orbivirus and Rotavirus, respectively. Ordered structures are visible in the periphery of the RNA region.
Wound tumor virus (WTV) has three protein shells, including an outer amorphous layer, an internal layer of distinct capsomers and a smooth core that is about 50 nm in diameter but lacking spikes.
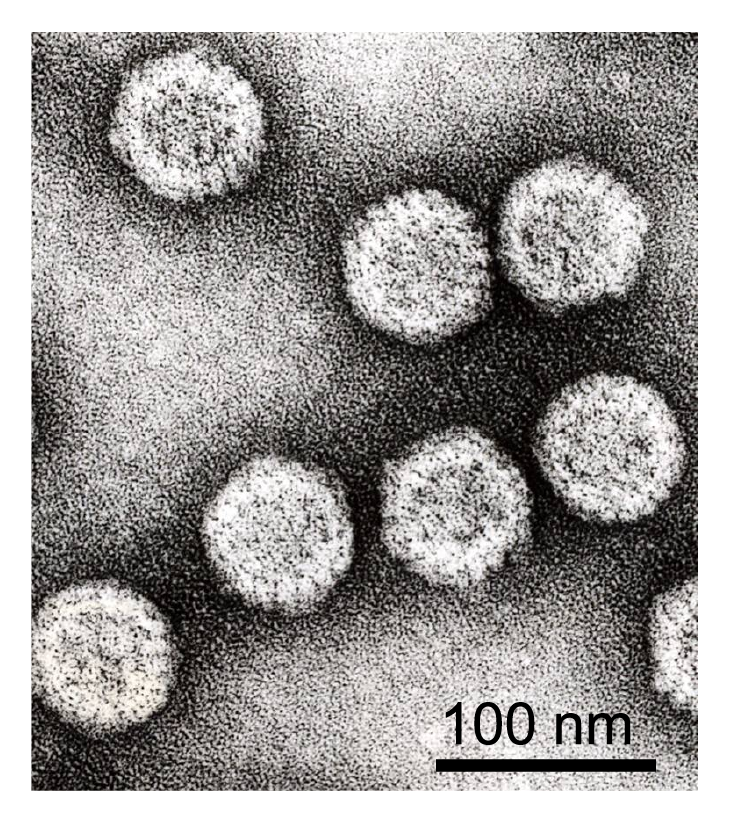 |
| Figure 1. Phytoreovirus Negative-contrast electron micrograph of rice gall dwarf virions (Courtesy of T. Wei). |
Physicochemical and physical properties
The Mr of phytoreoviruses is about 75×106. The virion S20,w is about 510. The optimal stability of particles is at pH 6.6. The buoyant density of RDV is 1.39–1.42 g cm−3 and the virion is unstable losing P8 in CsCl. CCl4 removes P2 from the RDV virion.
Nucleic acid
Phytoreoviruses have 12 genome segments of linear dsRNA, numbered according to their migration during polyacrylamide gel electrophoresis (PAGE). However, this order is misleading for the pairs Seg4/5 and Seg9/10, since sequence analysis indicates that Seg 5 and Seg10 are longer than their electrophoretic mobility would indicate (Table 1.Phytoreovirus).
Table 1.Phytoreovirus Genome segments and protein products of rice dwarf virus, Akita isolate
|
Genome segment |
bp |
Non-coding region lengths 5′ – 3′ (bp) |
Protein |
Protein mass (kDa)* |
Location and Function |
|||
|
Seg1 |
4423 |
35–53 |
P1 |
164.1 (170) |
Core, RNA-directed RNA polymerase |
|||
|
Seg2 |
3512 |
14–147 |
P2 |
123.0 (130) |
Minor outer capsid, essential for vector transmission |
|||
|
Seg3 |
3195 |
38–97 |
P3 |
114.3 (110) |
Core capsid |
|||
|
Seg4 |
2468 |
63–221 |
Pns4 |
79.8 (83) |
Non-structural, phosphorylated, forms minitubules |
|||
|
Seg5 |
2570 |
26–138 |
P5 (Cap) |
90.5 (89) |
Core, capping enzyme (guanylyltransferase and transmethylase) |
|||
|
Seg6 |
1699 |
48–121 |
Pns6 |
57.4 (56) |
Non-structural, movement protein, viroplasm matrix protein |
|||
|
Seg7 |
1696 |
25–150 |
P7 |
55.3 (58) |
Core, nucleic acid binding protein |
|||
|
Seg8 |
1427 |
23–138 |
P8 (T13) |
46.5 (43) |
Major outer capsid protein with T=13 symmetry |
|||
|
Seg9 |
1305 |
24–225 |
Pns9 |
38.9 (49) |
Minor outer capsid |
|||
|
Seg10 |
1321 |
26–233 |
Pns10 |
39.2 (35) |
Non-structural, silencing suppressor, forms tubular structure, essential for vector transmission |
|||
|
Seg11 |
1067 |
29–492 |
Pns11a |
20.0 (23) |
Non-structural, nucleic acid binding protein, silencing suppressor, viroplasm matrix protein |
|||
|
|
|
5–492 |
Pns11b |
20.8 (24) |
|
|||
|
Seg12 |
1066 |
41–86 |
Pns12 |
33.9 (34) |
Non-structural, phosphorylated viroplasm matrix protein |
|||
|
|
|
312–475 |
Pns12OPa |
|
|
|||
|
|
|
336–475 |
Pns12OPb |
9.6 (7) |
|
|||
* Calculated from nucleotide sequence (length determined by SDS PAGE in brackets).
RNA constitutes about 22% of the virion dry weight. The dsRNA Mr is in the range 0.3 to 3.0×106, with characteristic masses for different viruses. For WTV Seg4 = 2,565 bp; Seg5 = 2,613 bp; Seg6 = 1,700 bp; Seg7 = 1,726 bp; Seg8 = 1,472 bp; Seg9 = 1,382 bp; Seg10 = 1,172 bp; Seg11 = 1,128 bp and Seg12 = 851 bp. G+C content is 38–44% and 41–48% for the genomic segments of WTV and RDV respectively. The positive-sense strand of each genome segment, of all viruses in the genus, contains the conserved sequence 5′-GG(U/C)A---UGAU-3′ except for RDV Seg9 which has 5′-GGUA---CGAU-3′ (Table 2.Phytoreovirus). Internal to these genus-specific terminal sequences are inverted repeats of 6–14 bp. These repeat sequences differ for each RNA segment. Individual isolates of RDV can frequently be distinguished by their PAGE profile. RDV particles encapsidate the genomic dsRNA in supercoiled form.
Table 2.Phytoreovirus Conserved phytoreovirus terminal sequences (positive-sense strand)
|
Virus species |
Virus name |
5′-end |
3′-end |
|
Rice dwarf virus |
rice dwarf virus (RDV) |
5′-GGU/CAAA |
U/CGAU-3′ |
|
Rice gall dwarf virus |
rice gall dwarf virus (RGDV) |
5′-GGU/CAA/UUUU |
UGAU-3′ |
|
Wound tumor virus |
wound tumor virus (WTV) |
5′-GGUAUU |
UGAU-3′ |
|
(unclassified) |
Homalodisca vitripennis reovirus (HoVRV) |
5′-GGCG/A |
U/CGAU-3′ |
Proteins
Phytoreoviruses have six or seven structural proteins in the range 45 to 160 kDa. RDV has seven structural proteins (P1 (RdRP), P2, P3, P5 (Cap), P7, P8 and P9). Protein constitutes about 78% of the particle dry weight. Removal of the outer shell is not required for activation of the virus transcriptase and associated enzymes. Removal of RDV P2 abolishes the ability to infect vector cell monolayers but virions without P2 retain viral transcriptase activity and can infect vector insects by an injection method (Omura et al., 1998, Pu et al., 2011). P1 is the RdRP and binds to genomic dsRNA. P7 has non-specific nucleic acid binding activity. P3 binds to itself, P7 and P8. P7 binds to P1 and P8. P5 is probably a guanylyl transferase and has GTP, ATP and UTP binding activities. P3 and P8 form virus-like particles in transgenic rice plants. P8 interacts with rice glycolate oxidase, a typical enzyme of peroxisomes. P7 contains dsRNA-binding domains. The core capsid of RDV is composed of: P3, the major protein, which encloses P1, the RdRP; P5, a putative guanylyl transferase; and P7, a protein with nucleic acid-binding activity. The outer capsid of the virion is composed of: the P2 protein, which is involved in the ability of the virus to infect insect vector cells, the major outer capsid protein, P8, and the minor outer capsid protein, P9.
Genome organization and replication
The coding strand of each dsRNA has a single ORF, except for Seg11 and Seg12 of RDV (Table 1.Phytoreovirus), Seg9 of rice gall dwarf virus (RGDV) and Seg9 of WTV. RDV Seg11 has two in-frame initiation codons and has two overlapping ORFs. RDV Seg12, RGDV Seg9 and WTV Seg9 have a second, small, out-of-frame ORF embedded within the major ORF, but there is no evidence that this is expressed. Five structural and five NS WTV proteins have been assigned to genome segments. RDV Seg1 encodes the RdRP. Laboratory strains having internal deletions in some segments, but intact termini, replicate and compete favorably with wild-type virus, although the proteins expressed are aberrant, and the ability of the viruses to be transmitted by vectors may be lost. Virus replication occurs in the cytoplasm of infected cells in association with viroplasms. WTV and RGDV are confined to phloem tissues of the plant host, whereas RDV can also multiply elsewhere.
There are several non-structural proteins. Pns4 is phosphorylated and is associated with large cytoplasmic fibrils and forms novel minitubules in infected cells. Pns6 functions as a movement protein in plants and is the major component of viroplasm matrix. Pns10 is a viral silencing suppressor in plants, and the component of virus-containing tubules essential for virus spread via insect vectors (Chen et al., 2012). Pns11 is also a viral silencing suppressor in plants, and the major component of the viroplasm matrix. Pns12 is a phosphorylated protein that initates virolasm formation in virus-infected cells.
Biology
Plant hosts are either dicotyledonous (WTV) or graminaceous (RDV and RGDV). WTV was originally identified in northeastern USA in Agalliopsis novella (Say, 1830) leafhoppers, and was found in New Jersey, USA, in a single periwinkle (Catharanthus) plant set out as bait for mycoplasmas in a blueberry (Vaccinium) field. WTV is transmitted in a persistent-propagative manner by Agallia constricta (Van Duzee, 1894), A. quadripunctata (Provancher 1872), and A. novella. The experimental plant host range of WTV is wide and encompasses many dicotyledonous plants. The name of WTV derives from the fact that infected plants develop phloem-derived galls (tumors) at wound sites, notably at the emergence of side roots.
RDV and RGDV have narrow and overlapping host ranges. RDV causes severe disease in rice crops in South-East Asia, China, Japan and Korea, Nepal and the Philippines. RGDV has been reported in Thailand, Malaysia and China. RDV induces white flecks and streaks on leaves, with stunting and excessive production of side shoots. RDV is the only plant reovirus that is not limited to the phloem. Plants infected with RDV are stunted and fail to bear seeds. Since the virus is widespread among rice plants in southern China and other Asian countries, it is considered likely to be the cause of a significant overall reduction in rice production. RDV does not provoke enlargement or division of infected cells and does not induce galls, enations, or tumors. RGDV was found in a rice field in Thailand and induces stunting, shoot proliferation, a dark green color and enations in rice.
Phytoreoviruses induce no marked disease in their insect vectors. Virus replication occurs in the cytoplasm of infected cells in association with viroplasm. In the vector, there are no particular tissue tropisms. However, RDV induces abnormalities in fat body cells and mycetocytes. RDV and RGDV are all transmitted propagatively by cicadellid leafhoppers (N. cincticeps (Uhler 1896), N. nigropictus (Stål 1870), Recilia dorsalis Motschulsky, 1859, and some other Nephotettix spp). N. cincticeps is the most signifïcant vector for RDV. R. dorsalis is the most significant vector for RGDV. Virions are acquired from plants shortly after feeding. The latent period in leafhoppers is about 10–20 days. Thereafter, infected insects have a life-long ability to transmit virus to plants. Phytoreoviruses are also transmitted transovarially in their insect vectors. For example, RDV can exploit the ancient oocyte entry path of the obligate symbiont bacterium Candidatus Sulcia muelleri in its leafhopper vectors for transovarial transmission by binding to the bacterial envelope (Jia et al., 2017). Moreover, RGDV is transovarially transmitted in female leafhopper vectors via virus-containing tubules. RGDV also is paternally transmitted by the male leafhopper vectors along with sperm (Chen et al., 2012). Paternal virus transmission is more efficient than maternal transmission. Such paternal virus transmission scarcely affects the fittness of adult males or their offspring and plays a pivotal role in virus survival during seasons unfavorable for rice hosts in the field. Experimental data suggest that phytoreoviruses are not mechanically transmissible from plant to plant. No seed transmission has been reported. RDV and RGDV naturally infect rice and a few grass weeds of the graminaceous plants.
Antigenicity
Members of the three phytoreovirus species are antigenically distinct for epitopes on the outer surface. However, the inner surface epitopes of the capsid of RDV and RGDV cross-react.
Species demarcation criteria
In addition to the other general criteria used throughout the family, members of a species in the genus Phytoreovirus may be identified by:
- Sequence analysis: The seven structural proteins and the five nonstructural proteins of WTV, RDV and RGDV have significant levels (19–50%) of amino acid sequence identity.
- Amino acid identities of the major capsid protein P8 and core protein P5 are >80% within species and <56% between species (Moriyasu et al., 2007, Zhang et al., 2007) (Figure 2.Phytoreovirus).
- Cross-hybridization using conditions designed to detect >80% similarity.
- Host plant species; dicotyledons (WTV), or the family Graminae (RDV and RGDV).
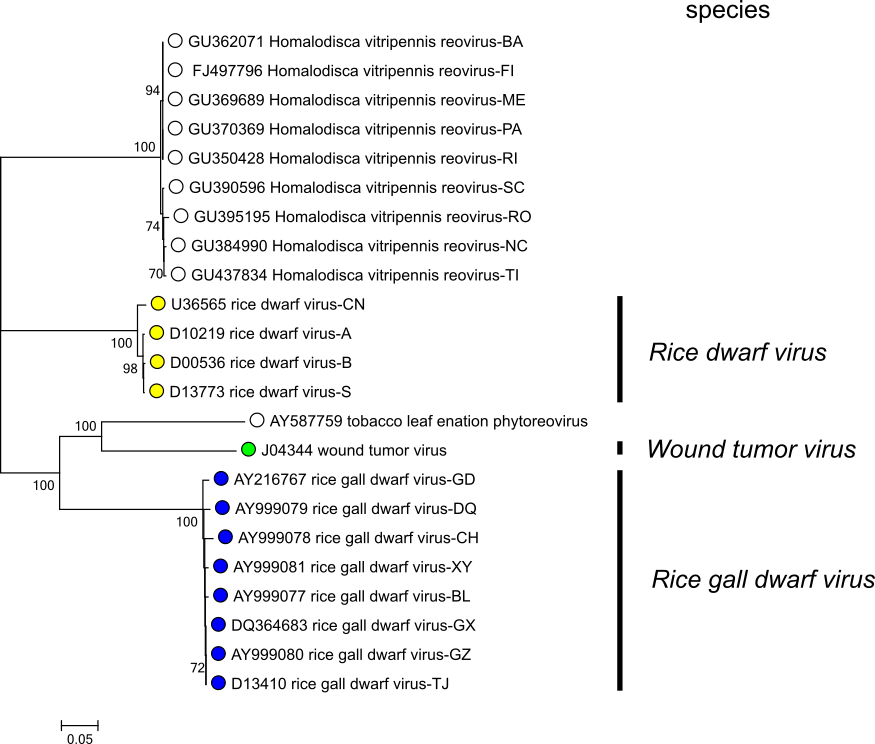 |
| Figure 2.Phytoreovirus Phylogenetic relationships between phytoreoviruses using the amino acid sequence encoded by Seg8. Sequences were aligned using MUSCLE (Edgar 2004) and the tree constructed in MEGA7 (Kumar et al., 2016) using the neighbour-joining method and the p-distance algorithm with pairwise deletion. Bootstrapping values (1000 replicates) above 70% are shown. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Related, unclassified viruses
|
Virus name |
Accession numbers |
Abbreviation |
|
Homalodisca vitripennis reovirus Bakersfield |
Seg1: GU362064; Seg2: GU362065; |
HoVRV-BA |
|
Homalodisca vitripennis reovirus Fillmore |
Seg1: FJ497789; Seg2: FJ497790; |
HoVRV-FI |
|
Homalodisca vitripennis reovirus isolate Mentone |
Seg1: GU369682; Seg2: GU369683; |
HoVRV-ME |
|
Homalodisca vitripennis reovirus isolate NC12 |
Seg1: GU384983; Seg2: GU384984; |
HoVRV-NC |
|
Homalodisca vitripennis reovirus isolate Pauma1 |
Seg1: GU370362; Seg2: GU370363; |
HoVRV-PA |
|
Homalodisca vitripennis reovirus isolate Riverside |
Seg1: GU350421; Seg2: GU350422; |
HoVRV-RI |
|
Homalodisca vitripennis reovirus isolate Robeson |
Seg1: GU395188; Seg2: GU395189; |
HoVRV-RO |
|
Homalodisca vitripennis reovirus isolate SC20 |
Seg1: GU390589; Seg2: GU390590; |
HoVRV-SC |
|
Homalodisca vitripennis reovirus isolate Tifton4 |
Seg1: GU437827; Seg2: GU437828; |
HoVRV-TI |
|
rice bunchy stunt virus |
RBSV |
|
|
tobacco leaf enation phytoreovirus |
Seg5: AY587757; Seg7: AY587758; |
TLEV |
Virus names and virus abbreviations are not official ICTV designations.
The vector for Homalodisca vitripennis reovirus is the glassy-winged sharpshooter (Homalodisca vitripennis (Germar, 1821)). The vector for tobacco leaf enation phytoreovirus is unknown. Rice bunchy stunt virus is a possible species of phytoreovius (Hibino 1996), and is transmitted in a persistent-propagative manner by N. cincticeps and N. virescens.

