Family: Spinareoviridae
Genus: Mycoreovirus
Distinguishing features
Virions have a relatively featureless outer capsid as viewed by negative-staining and electron microscopy, whereas the core particles have 12 icosahedrally arranged surface turrets or spikes (Hillman et al., 2004, Wei et al., 2003). The genome is composed of 11 or 12 segments of dsRNA. The members of the genus that have been described all infect fungi (Hillman and Suzuki 2004).
Virion
Morphology
Particles are non-enveloped. Electron microscopy and negative-staining of mycoreovirus virions with aqueous uranyl acetate indicates that they are double-layered, spherical in appearance (icosahedral symmetry) and approximately 80 nm in diameter. The viral core (estimated as 50 nm in diameter) has 12 icosahedrally arranged surface projections (turrets or B-spikes) (Figure 1 Mycoreovirus) (Wei et al., 2003). Particles are disrupted by 2% phosphotungstic acid (pH 7.0).
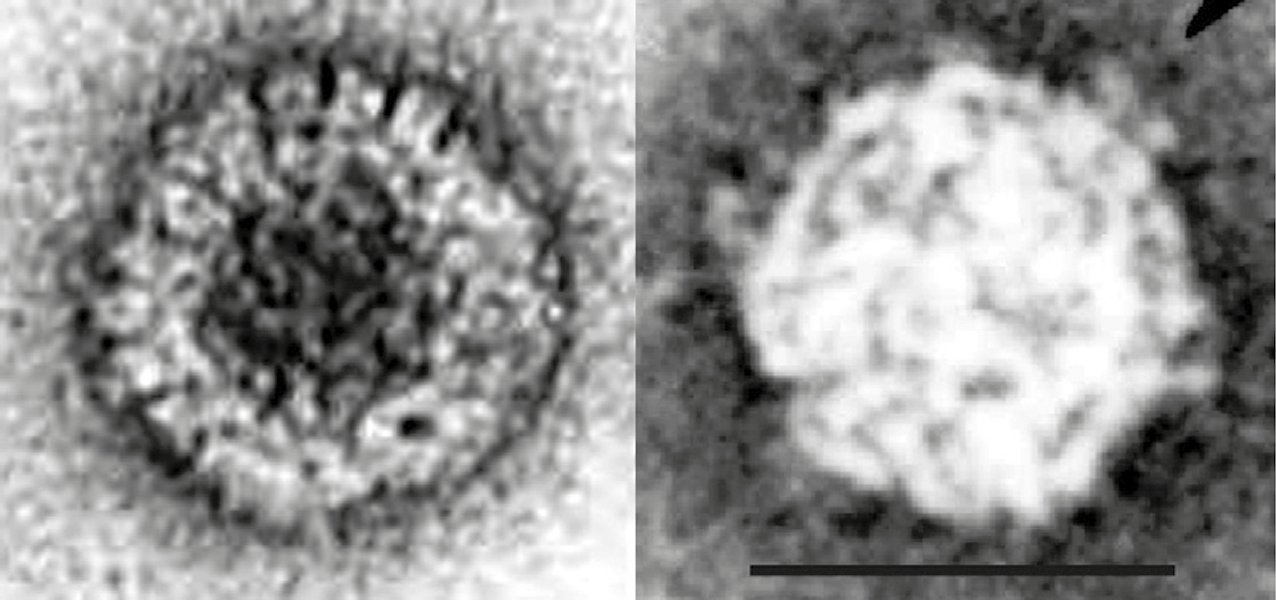 |
| Figure 1 Mycoreovirus (Left) Electron micrograph of virions of Cryphonectria parasitica mycoreovirus 1, after purification by sucrose gradient centrifugation, stained with 1% uranyl acetate (courtesy of B. Hillman). (Right) Core particle of Rosellinia necatrix mycoreovirus 3 possessing icosahedral arrangement surface projections (turrets or spikes), stained with 1% uranyl acetate (courtesy of C. Wei). The bar (right panel) represents 50 nm. |
Nucleic acid
The genome consists of 11 (Cryphonectria parasitica mycoreovirus 1, CpMYRV1 and Cryphonectria parasitica mycoreovirus 2, CpMYRV2; group 1) or 12 (Rosellinia necatrix mycoreovirus 3, RnMYRV3; group 2) dsRNA segments that are numbered in order of reducing molecular weight or increasing electrophoretic mobility during agarose gel electrophoresis (AGE). The genome of CpMYRV1 is 23,436 bp in total, with the length of individual segments ranging from 732 bp to 4,127 bp. Gel-electrophoresis of individual segments results in characteristic migration patterns. The CpMYRV1 genome shows a 3-3-2-3 electrophoretic profile following either 11% polyacrylamide gel electrophroresis (PAGE) or 1% AGE (Suzuki et al., 2004, Hillman et al., 2004). The genomic RNAs of RnMYRV3 have a 3-3-6 electrophoretic profile following 5% PAGE with segments ranging from 943 bp to 4,143 bp (Wei et al., 2004, Wei et al., 2003, Osaki et al., 2002). As with other members of the order Reovirales, the genome segment migration patterns during AGE (or low percentage (<5%) PAGE) are considered likely to be characteristic of each virus species. Note that RnMYRV3 undergoes spontaneous loss of one of the genomic segments, Seg8, during subculturing of infected fungal strains (Kanematsu et al., 2004). CpMYRV1 has no homolog of RnMYRV3 Seg8 (Suzuki et al., 2004). Terminal sequences of the genome segments are shown in Table 1 Mycoreovirus.
Table 1 Mycoreovirus Conserved mycoreovirus terminal sequences (positive-sense strand)
| Virus species | Virus name | 5′-end | 3′-end |
|---|---|---|---|
| Mycoreovirus alcryphonectriae | Chryphonnectria parasitica mycoreovirus 1 (CpMYRV1) | 5′-GAUCA | CGCAGUCA-3′ |
| Mycoreovirus roselliniae | Rosellinia necatrix mycoreovirus 3 (RnMYRV3) | 5′-ACAAUUU | UGCAGAC-3′ |
Proteins
The structural proteins have not been identified.
Genome organization and replication
The segment lengths and predicted masses of proteins encoded by the 11 segments of CpMYRV1 and the 12 segments of RnMYRV3 are shown in Table 2 Mycoreovirus and Table 3 Mycoreovirus, respectively. Proteins are named as VP1 to VP11 or VP12, based on the molecular weight of the genome segment (segment number) from which they are translated. Expression of CpMYRV1 proteins using a modified baculovirus identified the capping enzyme as VP3 by an autoguanylation assay(Supyani et al., 2007). Using a series of progressive N-terminal and C-terminal deletion mutants the auto-guanylation active-site of VP3 was localized to amino acid residues 133–667. Within this region, a sequence was identified (residues 170–250) that has relatively high sequence similarity to homologues in the mycoreoviruses Chryphonnectria parasitica mycoreovirus 2 (CpMYRV2) and RnMYRV3, and two coltiviruses, Colorado tick fever virus and Eyach virus. Site-directed mutagenesis of conserved residues revealed that H233, H242, Y243, F244 and F246, but not K172 or K202, play critical roles in guanylyltransferase activities. The CpMYRV1 Seg4-encoded protein, VP4, is not essential for viral replication but is required for efficient vertical transmission via conidia and for the induction of normal symptoms (Eusebio-Cope et al., 2010). VP10, encoded by CpMYRV1 Seg10, is also dispensable for CpMYRV1 replication, but contributes to virulence reduction and reduced growth of aerial mycelia (Eusebio-Cope et al., 2010, Sun and Suzuki 2008).
Table 2.Mycoreovirus Genome segments and protein products of Cryphonectria parasitica mycoreovirus 1
| Genome segment | bp | Protein nomenclature | Protein aa (mass, kDa) | Function [homologues] |
|---|---|---|---|---|
| Seg1 | 4127 | VP1 | 1354 (151.8) | RNA-directed RNA polymerase (RdRP, Pol); coltivirus VP1 |
| Seg2 | 3846 | VP2 | 1238 (138.5) | [coltivirus VP2] |
| Seg3 | 3251 | VP3 | 1065 (120.8) | Guanylyltransferase (Cap) |
| Seg4 | 2269 | VP4 | 721 (79.8) | [RnMYRV3-W370 Seg4 and coltivirus Seg4] |
| Seg5 | 2023 | VP5 | 648 (72.8) | |
| Seg6 | 2056 | VP6 | 650 (73.4) | [RnMYRV3-W370 Seg6 and coltivirus Seg10] |
| Seg7 | 1536 | VP7 | 482 (54.1) | |
| Seg8 | 1539 | VP8 | 470 (51.2) | |
| Seg9 | 1072 | VP9 | 298 (32.9) | [RnMYRV3-W370 Seg11] |
| Seg10 | 975 | VP10 | 248 (27.8 | |
| Seg11 | 732 | VP11 | 102 (11.5) |
Table 3 Mycoreovirus Genome segments and protein products of Rosellinia necatrix mycoreovirus 3
| Genome segment | bp | Protein nomenclature | Protein aa (mass, kDa) | [Homologue] |
|---|---|---|---|---|
| Seg1 | 4143 | VP1 | 1360 (153.4) | |
| Seg2 | 3773 | VP2 | 1226 (138.5) | [coltivirus VP2] |
| Seg3 | 3310 | VP3 | 1086 (121.9) | |
| Seg4 | 2259 | VP4 | 725 (78.7) | |
| Seg5 | 2089 | VP5 | 646 (72.3) | |
| Seg6 | 2030 | VP6 | 634 (71.5) | |
| Seg7 | 1509 | VP7 | 482 (55.1) | |
| Seg8 | 1299 | VP8 | 325 (36.5) | |
| Seg9 | 1226 | VP9 | 380 (41.6) | |
| Seg10 | 1171 | VP10 | 310 (33.6) | |
| Seg11 | 1003 | VP11 | 282 (31.1) | |
| Seg12 | 943 | VP12 | 265 (29.2) |
On the basis of the available sequence data for several of the genome segments and the overall similarity of mycoreoviruses to other members of the order Reovirales, it is assumed that many aspects of the genome organization and replication are also similar. On this basis, it is likely that the viral core contains transcriptase complexes that synthesize mRNA copies of the individual genome segments, which are exported and translated to produce viral proteins within the host cytoplasm. These positive-sense RNAs also are likely to form templates for negative-sense strand synthesis during progeny virion assembly and maturation. As in the case of other members of the order Reovirales, most of the mycoreovirus genome segments appear to represent single genes, with a large ORF and relatively short terminal non-coding regions. Many rearranged genome segments of CpMYRV1 are generated in RNA silencing-compromised fungal strains (Eusebio-Cope et al., 2010, Sun and Suzuki 2008, Tanaka et al., 2011, Eusebio-Cope and Suzuki 2015).
Biology
RnMYRV3 is found in the mycelium of a strain of the white root rot fungus Rosellinia necatrix. The virus itself appears to make the fungus hypovirulent and may represent a useful biological control for the damage caused by the wild-type fungus (Kanematsu et al., 2004). The uninfected fungus can be regenerated by hyphal tip culture. CpMYRV1 and CpMYRV2 are found in the mycelium of the filamentous fungus that causes chestnut blight disease (Cryphonectria parasitica) (Hillman and Suzuki 2004). Purified particles of CpMYRV1 can be used to infect protoplasts of virus-free mycelium. Infection with CpMYRV1 greatly reduces fungal virulence and may represent a useful biological control for the disease (Hillman et al., 2004). CpMYRV2 alters the colony morphology and reduces the virulence of C. parasitica (Hillman et al., 2004), although the infection is unstable (Aulia et al., 2019).
Species demarcation criteria
Members of different species are distinguished by the number of dsRNA segments, host range and degree of amino acid sequence identity. For example, although the viruses CpMYRV1 and CpMYRV2 both have 11 genome segments and infect C. parasitica, their capping enzymes (VP3) are only 29% identical, justifying their classification into the species Mycoreovirus alcryphonectriae and Mycoreovirus becryphonectriae (Supyani et al., 2007). Similarly, the highest amino acid identity between homologous proteins of RnMYRV3 and CpMYRV1 or CpMYRV2 is 39% for the RdRP of CpMYRV1 (Suzuki et al., 2004), which, together with its different host (Rosellinia necatrix) and number of segments (12), justify RnMYRV3 being classified as a member of a third species, Mycoreovirus roselliniae.
Members of both C. parasitica mycoreoviruses species (Mycoreovirus alcryphonectriae and Mycoreovirus becryphonectriae, CpMYRV1 and CpMYRV2) contain 11 genome segments (Group 1), but only have approximately 29% amino acid sequence identity in the capping enzyme, confirming their identity as members of distinct species (Supyani et al., 2007).
There are also some clear indications of homology in the longer genome segments of CpMYRV1 and CpMYRV2 mycoreoviruses to RnMYRV3, Group 2, 12 segment genome). The highest amino acid sequence identity was detected in the RdRP sequence (39% identity) (Suzuki et al., 2004).
Related, unclassified viruses
| Virus name | Accession number | Abbreviation |
| Sclerotinia sclerotiorum mycoreovirus 4 | Seg1: KU128375; Seg2: KU128376; Seg3: KU128377; Seg4: KU128378; Seg5: KU128379; Seg6: KU128380; Seg7: KU128381; Seg8: KU128382; Seg9: KU128383; Seg10: KU128384; Seg11: KU128385; Seg12: KU128386 | SsMYRV4 |
Virus names and virus abbreviations are not official ICTV designations.
Another mycoreovirus with an 12-segmented genome, Sclerotinia sclerotiorum mycoreovirus 4 (SsMYRV4), has been reported from an ascomycetous fungus (Sclerotinia sclerotiorum). This virus has a closer phylogenetic relationship to RnMYRV3 than to CpMYRV1 (Wu et al., 2017).

