Family: Spinareoviridae
Genus: Aquareovirus
Distinguishing features
Aquareoviruses physically resemble orthoreoviruses but possess 11 dsRNA genome segments. They infect a variety of aquatic animals, including finfish and crustaceans. Aquareoviruses replicate in cell cultures of piscine and mammalian origin, at temperatures from 15 and 28 °C. Large syncytia are produced as a typical cytopathic effect of infection by a majority of aquareoviruses.
The highest level of amino acid sequence identity detected between the RdRP of aquareoviruses and a member of a distinct reovirus genus is 41% (to mammalian orthoreovirus 3 (MRV3), a member of the genus Orthoreovirus), supporting the hypothesis that these genera are closely related (derived from a common ancestor, estimated ca. 510 million years ago). Although this value of amino acid sequence identity is higher than that separating most genera (usually <30%), classification of the aquareoviruses and orthoreoviruses as members of two distinct genera is based on multiple parameters and not simply genetic relatedness. For example, aquareoviruses can infect members of many marine and freshwater species of fish, whereas orthoreoviruses primarily infect mammals, birds and reptiles, suggesting co-speciation of the viruses with their respective hosts.
Virion
Morphology
Aquareovirus particles are spherical in appearance with a diameter of about 80 nm and composed of multiple capsid layers (Figure 1 Aquareovirus, upper and middle panels). The outermost layer, formed by VP5-VP7 heterodimers, consists of 600 subunits (200 trimers) arranged on an incomplete T=13 icosahedral lattice, with an overall structural organization identical to those of mammalian orthoreovirus 3 (MRV3) and avian orthoreovirus (ARV). A distinguishing feature on the outer layer is the five-fold proximal depressions, resulting from missing peripentonal trimers (Figure 1 Aquareovirus, upper left panel, P1 position indicated by arrows). The shaded surface view of the aquareovirus core structure (Figure 1 Aquareovirus, upper right panel), shows the innermost capsid shell, which is about 60 nm in diameter. Twelve VP1 pentameric turrets decorate the shell of 120 VP3 monomers, which are arranged with icosahedral symmetry that is interpreted as T=1 (comparable to the sub-core of the orbiviruses and the innermost capsid shell of the rotaviruses) and are clamped together by 120 VP6 monomers.
Removal of VP7 generates infectious subviral particles (ISVPs), which have a smooth surface formed by a network of VP5 trimers (Figure 1 Aquareovirus, bottom panel). The atomic model of ISVPs contains six conformers of four proteins: two of VP3, two of VP6, one VP1 on the core, and one VP5 on the coat.
The particle morphology of aquareoviruses is strikingly similar to that of orthoreovirus ISVPs. A noticeable morphological distinction between aquareovirus and orthoreoviruses is that aquareovirus particles lack the hemagglutinin spike protein σ1 observed on orthoreovirus particles.
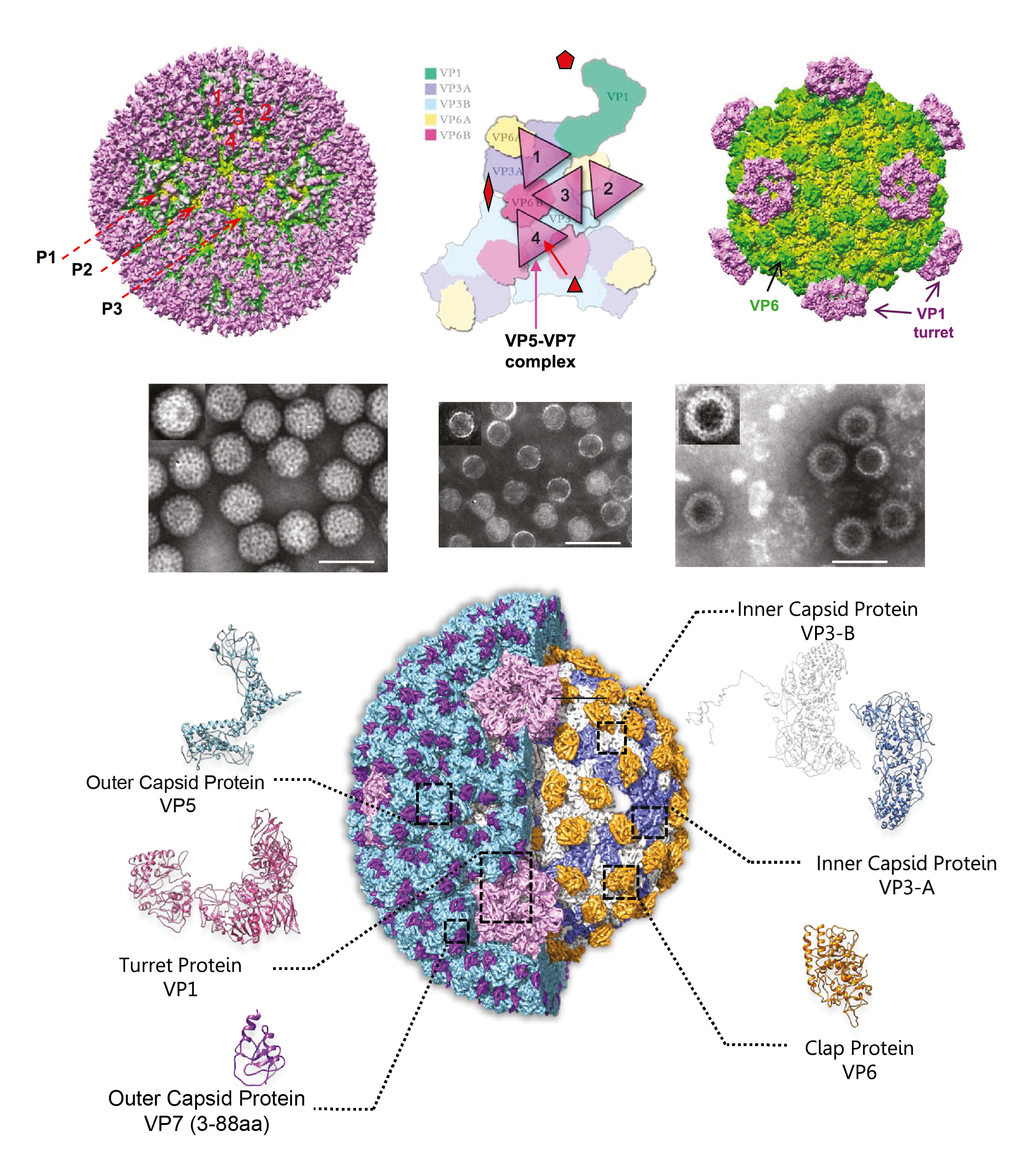 |
| Figure 1. Aquareovirus Aquareovirus particle structure. (Top panel) Structural representation of grass carp reovirus (GCRV; Aquareovirus ctenopharyngodontis) virion (left) and core (right) by cryoEM. Triangles (middle) represent the VP5–VP7 complex on the virion (from (Cheng et al., 2008)). (Middle panel) Transmission electronmicroscopy (TEM) of negatively stained GCRV particles. From left to right: intact virion, core and top components (empty particles). The scale bar represents 100 nm (From (Fang et al., 2008)). (Bottom panel) Complete atomic model of native degraded particle or infectious/intermediate subviral particle (ISVP) of GCRV. On the right-hand side, it shows the core frame proteins after removal of the outer coat proteins VP5 and VP7. Ribbon models of the atomic structures of the six conformers from five structural proteins are shown in the periphery. The residues 3–88 amino acid (aa) of VP7 were resolved (Courtesy of Q. Fang). |
Physicochemical and physical properties
The virion buoyant density in CsCl is 1.36 g cm−3 with a sedimentation coefficient of about 550S. Virion infectivity is stable between pH 3 and pH 10. Virion infectivity is not affected by treatment with ether or chloroform. Exposure to UV irradiation reduces infectivity. None of the viral proteins are removed from the particle by treatment with 3 mM EDTA or cesium salts. Aquareoviruses held at 4 °C, 16 °C or 23 °C in minimal essential medium (MEM) with 5% serum show no significant reduction in infectivity over a period of 28 days. However, all virion infectivity is lost after incubation at 45 °C for 7 days. Virion infectivity is rapidly inactivated by heating to 56 °C.
Nucleic acid
The aquareovirus genome is composed of 11 segments of dsRNA that are packaged in equimolar ratios. The Mr of the dsRNA segments range from 0.4 to 2.6×106. The total Mr of the golden shiner reovirus (GSRV) is about 1.5×107 (23,695 bp). The genomic RNAs are named segment 1 (Seg1) to segment 11 (Seg11) in order of increasing electrophoretic mobility in 1% agarose gels. The genome segments migrate as three length classes. There are three long (Seg1 to Seg3, about 3.9–3.8 kbp), three medium (Seg4 to Seg6, about 2.3–2.0 kbp) and five short segments (Seg7 to Seg11, about 1.4–0.8 kbp). Members of six distinct species (Aquareovirus salmonis, Aquareovirus oncorhynchi, Aquareovirus ctenopharyngodontis, Aquareovirus ictaluri, Aquareovirus scophthalmi and Aquareovirus maculosi) were originally delineated by reciprocal RNA–RNA hybridization studies but can also be distinguished by nucleotide sequence analyses. The genome segment migration pattern (electropherotype), as analyzed by electrophoresis in 1% agarose gel, is consistent within a single species but shows significant variation among species. However, viruses within a species can have variations in electropherotype when their dsRNA genome segments are analyzed by electrophoresis in high percentage (>6%) polyacrylamide gels.
The G+C content of aquareovirus RNA ranges between 52 and 60%. The complete genomic sequences of GSRV and grass carp reovirus (GCRV) have been described from cloned cDNAs, as have individual genome segments of other aquareovirus isolates. Genomic dsRNA segments have 7 bp at the 5′-terminus and 6 bp at the 3′-terminus that are conserved in all 11 genome segments for members of a particular virus species (Table 1 Aquareovirus).
Table 1 Aquareovirus Conserved terminal aquareovirus sequences (positive-sense strand)
| Virus species | Virus names | 5′-end | 3′-end |
|---|---|---|---|
| Aquareovirus salmonis | chum salmon reovirus (CHSRV) | 5′-GUUUUAU/G | A/UUCAUC-3′ |
| Aquareovirus ctenopharyngodontis | golden shiner reovirus (GSRV) grass carp reovirus (GCRV) | 5′-GUUAUUU/G | A/UUCAUC-3′ |
| Aquareovirus graminis | American grass carp reovirus (AGCRV) | 5′-GUUUUAU/A | U/AU/AUCAUC-3′ |
Proteins
Chum salmon reovirus isolates contain seven structural proteins: VP1, 130 kDa; VP2, 127 kDa; VP3, 126 kDa; VP4, 73 kDa; VP5, 71 kDa; VP6, 46 kDa; VP7, 35 kDa. The proteins VP1, VP2, VP3, VP4 and VP6 form the core of the virion, with VP3 and VP6 more abundant than VP1 and VP2. VP6 and VP3 probably form nodules and the spherical shell of the core, respectively. VP1 is thought to form turret-like structures present at the five-fold axis. VP2 is present in very small amounts per virion and is thought to be present beneath the five-fold axis.
VP7 and VP5 are present in the outer coat of the virion. Both proteins are removed by prolonged trypsinization, resulting in release of core particles. VP7 is the most external protein. VP5 is the next most accessible protein after VP7. Removal of VP7 by trypsin may expose some regions of VP5 critical for efficient entry into cells.
Lipids
Aquareovirus particles have no known lipid components.
Carbohydrates
VP7 of Aquareovirus salmonis isolates may be glycosylated.
Genome organization and replication
Twelve primary gene products are encoded by isolates of Aquareovirus salmonis
(Table 2 Aquareovirus). However, observed variations in dsRNA electropherotypes suggest that virions from other aquareovirus species may have proteins with significant differences in mass. Each genome segment of Aquareovirus salmonis isolates encodes only one primary translation product, with the exception of Seg11, which encodes two primary translation products. In addition to the seven structural proteins, the genome encodes five non-structural proteins of unknown function. In isolates of Aquareovirus ctenopharyngodontis and Aquareovirus graminis, it is Seg7 that encodes two proteins, from non-overlapping and out-of-phase ORFs (Figure 2 Aquareovirus).
Table 2 Aquareovirus Genome segments and protein products of striped bass reovirus (SBRV) (species Aquareovirus salmonis)
| Genome segment | kbp | Protein nomenclature | Protein mass (kDa) | Protein location | Function |
|---|---|---|---|---|---|
| Seg1 | 3.8 | VP1 | 130 | Inner capsid (core), forms pentameric turrets on the outer surface of the innermost capsid shell at fivefold vertices. | Guanylyltransferase and methylases |
| Seg2 | 3.6 | VP2 | 127 | Inner capsid (core), on the inner surface of the innermost capsid shell under fivefold vertices | RNA-directed RNA polymerase (RdRP) |
| Seg3 | 3.3 | VP3 | 126 | Inner capsid (core) | Innermost capsid shell |
| Seg4 | 2.5 | NS1 | 97 | Non-structural | Binds ssRNA and forms viral inclusion bodies |
| Seg5 | 2.4 | VP5 | 71 | Inner capsid (core) | NTPase |
| Seg6 | 2.2 | VP4 | 73 | Inner capsid (core), outer capsid protein | Membrane penetration protein |
| Seg7 | 1.5 | NS4 | 28 | Non-structural | Induces cell–cell fusion |
| Seg8 | 1.4 | VP6 | 46 | Inner capsid (core) | Clamper protein, stabilizes innermost capsid shell |
| Seg9 | 1.2 | NS2 | 39 | Non-structural | ssRNA-binding |
| Seg10 | 0.9 | VP7 | 34 | Major outer capsid | Outermost protection protein |
| Seg11 | 0.8 | NS3 | 29 | Non-structural | unknown |
| NS5 | 15 | Non-structural | unknown |
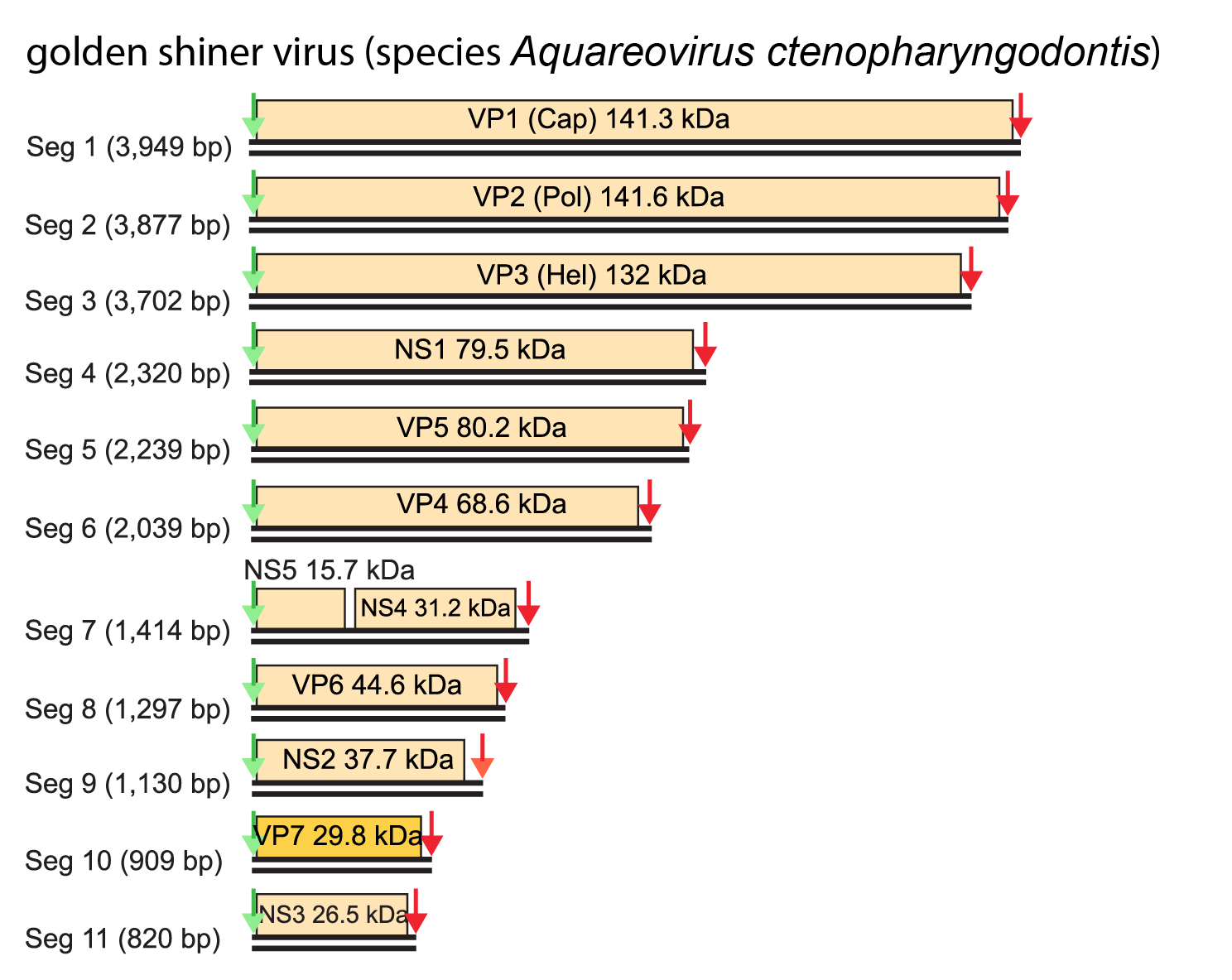 |
| Figure 2 Aquareovirus Genome organization of the 11 dsRNA segments of golden shiner virus (Aquareovirus ctenopharyngodontis). Each segment has a single ORF, except Seg7 which contains two ORFs. The green arrows indicate the upstream conserved terminal sequence (positive-sense strand 5′-GUUAUUU/G….) while the red arrows indicate the downstream conserved terminal sequence (positive-sense strand ….A/UUCAUC-3′). |
Biology
Host range
Aquareoviruses have been isolated from poikilothermic vertebrates as well as invertebrates (hosts include fish and molluscs) obtained from both fresh and sea water. The viruses replicate efficiently in fish and mammalian cell lines at temperatures ranging from 15 °C to 28 °C. Aquareovirues produce a characteristic cytopathic effect consisting of large syncytia, and are generally of low pathogenicity in their host species. However, GCRV is highly pathogenic in grass carp. The infectivity of aquareoviruses is enhanced by treatment with trypsin or chymotrypsin, which correlates with digestion of the outer capsid protein VP7. The most infectious stage of the virus is produced by a 5-minute treatment with trypsin. However, prolonged trypsin treatment almost completely abolishes infectivity, reflecting release of core particles.
Antigenicity
Aquareovirus outer capsid proteins (CPs) lack hemagglutinating activity. Virions possess species and subspecies-specific antigenic determinants. Within a species, members may be antigenically related whereas members of different species are antigenically distinct. Minor antigenic cross-reactivity has only been demonstrated between members of Aquareovirus salmonis and Aquareovirus oncorhynchi. Distinct serotypes probably exist within each species. The major outer CP of members of Aquareovirus salmonis (VP7) is not the major neutralizing antigen. There is no antigenic relationship between aquareoviruses and mammalian orthoreoviruses.
Species demarcation criteria
In addition to the other general criteria used throughout the family, members of a species in the genus Aquareovirus may be identified by:
- Northern hybridization assays under conditions (stringency) that do not allow >17% mismatch. Only isolates within the same species will show hybridization.
- Sequence analysis: In genome segment 10 which encodes the major outer CP (VP7), viruses from different species have <55% nucleotide identity (<36% VP7 amino acid identity. In the RdRP, isolates of the same species have >95% aa identity, whereas the corresponding values between members of different species are 57–74%.
Related, unclassified viruses
| Virus name | Accession numbers | Abbreviation |
| chub reovirus* | CHRV | |
| golden ide reovirus | Seg2: AF450323; Seg5: AF450324 | GIRV |
| grass carp reovirus | Seg1: GQ896334; Seg2: GQ896335; Seg3: GU350742; Seg4: GU350743; Seg5: GQ896336; Seg6: GQ896337; Seg7: GU350744; Seg8: GU350745; Seg9: GU350746; Seg10: GU350747; Seg11: GU350748 | GCRV-HZ08 |
| grass carp reovirus | Seg1: HQ231198; Seg2: HQ231199; Seg3: HQ231200; Seg4: HQ231201; Seg5: HQ231202; Seg6: HQ231208; Seg7: HQ231203; Seg8: HQ231204; Seg9: HQ231205; Seg10: HQ231206; Seg11: HQ231207 | GCRVGD108 |
| grass carp reovirus | Seg1: JN967629; Seg2: JN967630; Seg3: JN967631; Seg4: JN967632; Seg5: JN967633; Seg6: JN967634; Seg7: JN967635; Seg8: JN967636; Seg9: JN967637; Seg10: JN967638; Seg11: JN967639 | GCRV104 |
| grass carp reovirus | Seg1: KC201177; Seg2: KC201178; Seg3: KC201179; Seg4: KC201180; Seg5: KC201181; Seg6: KC201182; Seg7: KC201183; Seg8: KC201184; Seg9: KC201185; Seg10: KC201186; Seg11: KC201187 | GCRV918 |
| hard clam reovirus* | HCRV | |
| landlocked salmon reovirus* | LSRV | |
| tench reovirus* | TNRV |
Virus names and virus abbreviations, are not official ICTV designations.
* See (Lupiani et al., 1995)
The sequences of isolates GCRV104, GCRV918, GCRVHZ08 and GCRVGD108 form a basally branching monophyletic group, distantly related to members of the genera Aquareovirus and Orthoreovirus (Wang et al., 2012, Ye et al., 2012, Pei et al., 2014, Crane and Carlile 2008).

