Family: Spinareoviridae
Genus: Idnoreovirus
Distinguishing features
Idnoreovirions have a roughly spherical outer capsid, which may have small icosahedrally-arranged surface projections when viewed by negative-staining and electron microscopy. The core particles have 12 icosahedrally-arranged surface turrets or spikes, which appear similar to those of cypovirions, but unlike cypoviruses, polyhedra are not formed and virions have a clearly defined outer capsid layer. The genome is composed of 10 segments of linear dsRNA. There is no evidence of significant sequence homology between idnoreovirus genes and those of other members of the order Reovirales, other than that of the RNA-directed RNA polymerase (RdRP) which has 20–27% amino acid sequence identity to the RdRP of viruses in other genera of the family Spinareoviridae. All members of the genus infect insects.
Virion
Morphology
Particles are non-enveloped with a clearly defined outer capsid layer (Figure 1 Idnoreovirus). Electron microscopy and negative-staining of virions (e.g., with aqueous uranyl acetate) shows that they are double-layered, roughly spherical in appearance (with icosahedral symmetry), with an estimated diameter of about 70 nm. Core particles (estimated diameter of ca. 60 nm) display 12 icosahedrally-arranged, prominent surface projections (ca. 15 nm in length – identified as turrets or spikes), which may lose a portion near the tip and at least in some cases appear to be tubular. In particles where stain has entered the central space, there appears to be material (considered likely to be protein) associated with the spike structure on the inside of the inner capsid shell (Figure 1 Idnoreovirus, far-right panel).
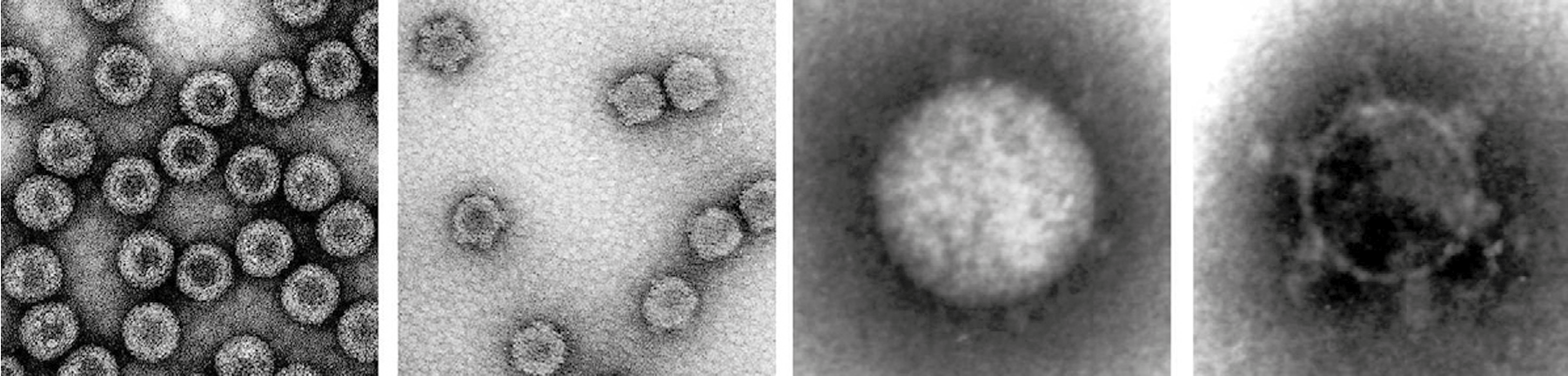 |
| Figure 1 Idnoreovirus Electron micrographs of purified virions (far left) and core particles (second left) of Hyposoter exiguae idnoreovirus 2, stained with uranyl acetate (courtesy of A. Makkay and D. Stoltz). Electron micrographs of a virion (second right) and core particle (far right) from purified preparations of Dacus oleae idnoreovirus 4 (DoIRV4), stained with sodium phosphotungstate (courtesy of M. Bergoin). DoIRV 4 virions have small icosahedrally arranged surface projections (estimated 12 in number). The DoIRV 4 cores have twelve large icosahedrally arranged spikes or turrets, which (like those of the cypoviruses) may lose a portion near to the tip. |
Physicochemical and physical properties
Limited studies of some viruses in the genus indicate that virions are resistant to freon (trichlorotrifluoroethane), CsCl and possibly chymotrypsin. Intact particles and cores of Diadromus pulchellus idnoreovirus 1 (DpIRV1) have densities of 1.370 g cm−3 and 1.385 g cm−3, respectively, and intact virions and empty particles of Dacus oleae idnoreovirus 4 (DoIRV4) have a density of about 1.38 g cm−3 and 1.28 g cm−3, respectively, as determined by CsCl gradient centrifugation. The outer capsid layer of Hyposoter exiguae idnoreovirus 2 (HeIRV2) can be disrupted by brief exposure to 0.4% sodium sarcosinate, releasing the virion core.
Nucleic acid
The genome usually consists of 10 dsRNA segments that are numbered in order of reducing molecular weight (or increasing electrophoretic mobility) during agarose gel electrophoresis (AGE). By analogy with other members of the order Reovirales, the genome segment migration patterns during AGE are likely to be characteristic for each idnoreovirus.
The total genome of DpIRV1 contains an estimated 25.15 kbp of dsRNA, with individual segments that range from about 4.8 to 0.98 kbp, showing a 5-5 electrophoretic migration pattern by 1% AGE. However, the virions of DpIRV1 may be unusual in the genus, since they can sometimes also contain an eleventh, 3.33 kbp dsRNA segment, the presence of which is related to, and may help determine, the sex and ploidy of the individual wasp host. This additional dsRNA (migrating between Seg3 and Seg4) contains sequences similar to, and therefore possibly derived by duplication of, an incomplete Seg3 (3.8 kbp).
The genome segments of HeIRV2 range in estimated length from about 3.9 to 1.35 kbp, with a 4–6 electrophoretic migration pattern by 12.5% polyacrylamide gel electrophoresis (PAGE). DoIRV-4 contains an estimated 23.4 kbp of dsRNA, with the estimated lengths of individual segments ranging from about 3.8 to 0.7 kbp and a 5-3-2 electrophoretic migration pattern by 7% PAGE. Ceratitis capitata idnoreovirus 5 (CcIRV5) has a 3-3-4 genome segment migration pattern by 6% PAGE, and has clear similarities to Drosophila melanogaster idnoreovirus 5 (DmIRV5), as analyzed by 0.5% agarose-2% polyacrylamide gels, suggesting that, despite some serological differences, these viruses belong to the same species (Idnoreovirus ceratitidis). It is unclear how closely Drosophila S virus (which causes the “S” phenotype in Drosophila simulans Sturtevant, 1919) is related to other Drosophila-derived idnoreoviruses. It is therefore currently listed as a related, unclassified member of the genus.
Another unclassified virus is Operophtera brumata idnoreovirus (ObIRV) with a genome of 23,647 kbp, with genome segments that range in length from 4.17 to 1.51 kbp, giving an electrophoretic migration pattern of 4-2-4. All 10 genome segments of ObIRV contain five fully-conserved bases at the 3′-termini of their positive-sense RNA strands, which are different from those found in other species that have been characterized within the order Reovirales. There is also conservation of four of the six 5′-terminal bases. In each case, the two terminal bases are complementary (5′-AA to 3′-TT).
In contrast, initial sequencing studies suggest that the 3′-termini of DpIRV1 genome segments are more variable than those of members of other species in the order Reovirales, with little sign of conservation. However, conserved sequences were detected at the 5′-termini (Table 1 Idnoreovirus), and these are different from those found in other reoviruses. No sequence data are currently available for other members of this genus.
Table 1 Idnoreovirus Conserved idneoreovirus terminal sequences (positive-sense strand)
| Virus species | Virus names | 5′-end | 3′-end |
|---|---|---|---|
| Idnoreovirus diadromi | Diadromus pulchellus idnoreovirus 1 (DpIRV1) | 5′-(a/u/g)CAAUUU | (variable)-3′ |
| unclassified | Operophtera brumata idnoreovirus (ObIRV) | 5′-AA(A/C)(A/U)AA | AGGUU-3′ |
Proteins
Structural proteins remain uncharacteristed.
Lipids
None reported.
Carbohydrates
Three proteins from DpIRV1 appeared to be glycosylated (ca. 21, 15 and 35 kDa).
Genome organization and replication
Native proteins of idnoreoviruses have not been characterized extensively. There is evidence that the genome of DpIRV1 has been subject to recombination and so details of genome organisation are instead given for ObIRV, an unclassified member of the genus. The ObIRV genome indicates that it encodes a total of 10 proteins, ranging in mass from 49.6 to 155.7 kDa and named VP1 to VP10 based on the molecular weight of the genome segment (segment number) from which they are translated.
The genome of DpIRV1 encodes 11 proteins with masses ranging from 21 to 140 kDa. On the basis of the overall similarity of idnoreoviruses to other members of the order Reovirales, it is assumed that many aspects of the genome organization and replication are similar. Thus, it is likely that the virus core contains transcriptase complexes that synthesize mRNA copies of the individual genome segments. These mRNAs are likely to be exported and translated to produce viral proteins in the host cytoplasm. These positive-sense RNAs are also likely to form templates for negative-sense strand synthesis during progeny virion assembly and maturation. Each of the genome segments that has been sequenced represents a single gene, with a single large ORF and relatively short terminal non-coding regions (NCRs) (Table 1 Idnoreovirus and Table 2 Idnoreovirus).
Table 2 Idnoreovirus Genome segments and protein products of Operophtera brumata idnoreovirus
| Genome segment | bp | Protein nomenclature | Protein aa (mass, kDa) | Structure/putative function |
|---|---|---|---|---|
| Seg1 | 4170 | VP1 | 1358 (155.7) | RNA-directed RNA polymerase |
| Seg2 | 3780 | VP2 | 1207 (137) | |
| Seg3 | 3595 | VP3 | 1161 (133) | T2 subcore shell |
| Seg4 | 3362 | VP4 | 1091 (122.7) | |
| Seg5 | 2106 | VP5 | 620 (69.2) | |
| Seg6 | 1935 | VP6 | 594 (68.4) | |
| Seg7 | 1606 | VP7 | 499 (57.1) | |
| Seg8 | 1584 | VP8 | 467 (51.6) | |
| Seg9 | 1547 | VP9 | 437 (49.6) | |
| Seg10 | 1509 | VP10 | 467 (53.9) |
Biology
Idnoreoviruses infect insects, in many cases with few pathological effects. However, they may significantly alter the biological properties of the host. Drosophila S virus appears to be associated with the S phenotype in D. simulans. The presence of an additional 3.33 kbp dsRNA segment in DpIRV1 is related to, and may help determine, the sex and ploidy of the host. This segment may play a role in the biology of this wasp species, possibly by providing information necessary for larval development.
Species demarcation criteria
See the general criteria used throughout the family as described on the family page
Related, unclassified viruses
| Virus name | Accession number | Abbreviation |
| Drosophila S virus | DSV | |
| Operophtera brumata idnoreovirus | Seg1: DQ192235; Seg2: DQ192236; Seg3: DQ192237; Seg4: DQ192238; Seg5: DQ192239; Seg6: DQ192240; Seg7: DQ192241; Seg8: DQ192242; Seg9: DQ192243; Seg10: DQ192244 | ObIRV |
Virus names and virus abbreviations are not official ICTV designations.

