Family: Sedoreoviridae
Genus: Orbivirus
Distinguishing features
Virions have a relatively featureless outer capsid as viewed by negative-staining and electron microscopy and a genome composed of 10 segments of dsRNA. Core particles have characteristic ring-shaped capsomers. Replication is accompanied by production of viral tubules and viral inclusion bodies (VIB) and may be accompanied by formation of flat hexagonal crystals/arrays of the major outer core protein, although these may be ‘rolled up’ to appear as ‘rods’ within the cytoplasm of infected cells. Viruses are transmitted between vertebrate hosts by a variety of hematophagous arthropods.
Virion
Morphology
Virions of bluetongue virus (BTV) are approximately 90 nm in diameter. Core particles have a maximum diameter of 73 nm, and sub-cores have a maximum diameter of 59 nm and an internal diameter of 46 nm (Figure 1.Orbivirus). The virion is spherical in appearance but has icosahedral symmetry. Although mature virions lack a lipid envelope, they can leave the host cell by budding through the plasma membrane. During this process, they transiently acquire a membrane envelope, which is usually regarded as unstable, although its functional significance has not been fully explored. Unpurified virions are often strongly associated with cellular membranes. By conventional electron microscopy, the surface of intact non-enveloped virions is indistinct (Figure 1.Orbivirus). However, the outer capsid does have an ordered structure, with icosahedral symmetry and sail-shaped surface projections that can be observed on virions where the particle structure is maintained (e.g., using cryoEM; Figure 2.Orbivirus). When the outer capsid layer is removed, it is possible to view the surface layer of the core particle, which is composed entirely of capsomeres of VP7 (T=13), with 780 copies arranged as 260 trimers in hexameric rings (pentameric at the five-fold axes; Figure 1.Orbivirus and Figure 2.Orbivirus). These rings, which are readily observed by conventional electron microscopy, give rise to the name of this genus. The core particle also contains a complete inner-capsid shell (the subcore layer), composed of 120 copies of VP3, which surrounds the 10 dsRNA genome segments. Three minor core proteins (the transcriptase complexes) are attached to the inner surface of the subcore at the 5-fold symmetry axes (Figure 1.Orbivirus). Assembly of the subcore layer appears to control the overall assembly, size and symmetry of the particle.
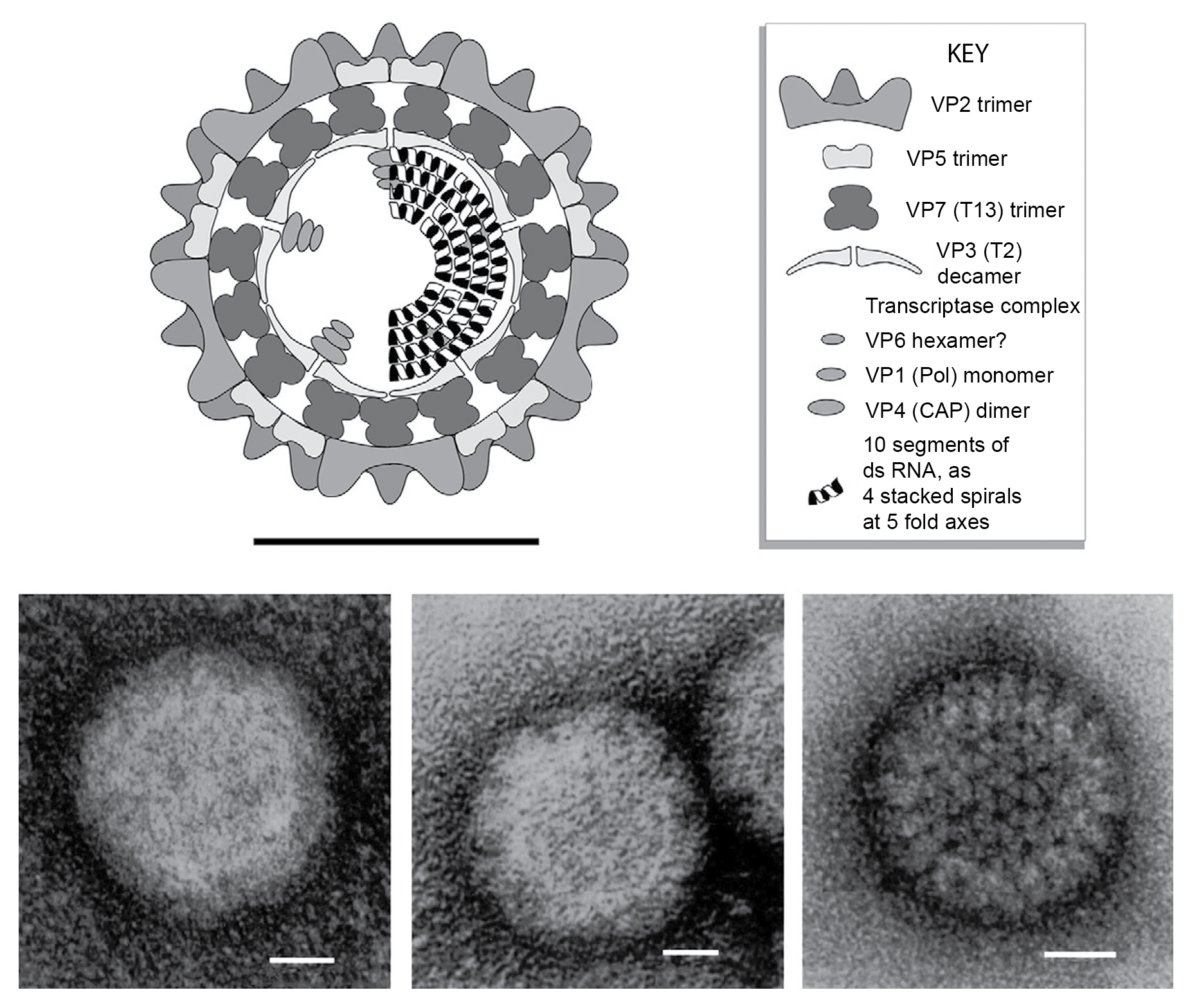 |
|
Figure 1. Orbivirus (Top) Diagram of bluetongue virus virion structure, constructed using data from biochemical analyses, electron microscopy, cryoEM and X-ray crystallography (courtesy of P.P.C. Mertens and S. Archibald). (Bottom) Electron micrographs of African horse sickness virus serotype 9 particles stained with 2% aqueous uranyl acetate. (Left) virions, having the relatively featureless surface structure. (Center) Infectious subviral particles, containing chymotrypsin cleaved outer capsid protein VP2 and showing some discontinuities in the outer capsid layer. (Right) core particles, from which the entire outer capsid has been removed, to reveal the structure of the VP7 (T13) core surface layer and showing the ring shaped capsomeres. (Courtesy of P. P. C. Mertens). |
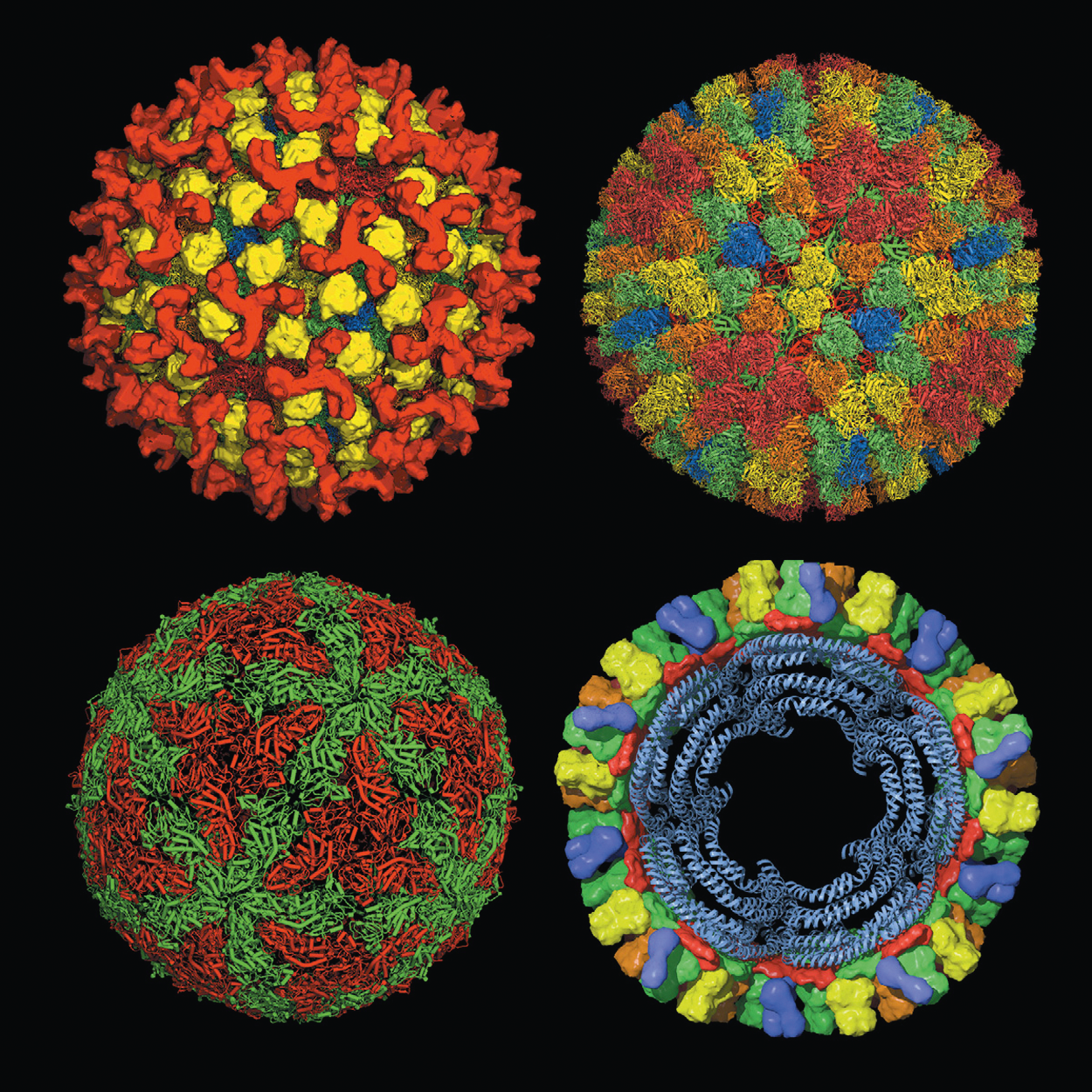 |
| Figure 2. Orbivirus (Top left) The outer capsid layer of bluetongue virus virions (from cryoEM) showing trimers of VP2 in red and trimers of VP5 in yellow, superimposed on the underlying X-ray crystallography structure for the virion core. (Top right) The structure of the virion core as determined by X-ray crystallography of the native core particle. The outer core surface, composed of 260 trimers of VP7 arranged with T=13 symmetry. The chemically identical but structurally different trimers are named and colored in order of increasing distance from the five-fold axes of symmetry (P [red], Q [orange], R [green], S [yellow] and T [blue] situated at the three-fold axes). (Bottom left) The virion subcore shell (from X-ray crystallography) is composed of 120 copies of VP3, arranged with T=2 symmetry. The chemically identical but structurally different molecules are shown: “A” (green: surrounding the five-fold axis) and “B” (red: surrounding the three-fold axis). (Bottom right) Model cross-section of the virion core showing packaging of the dsRNA as four concentric shells. (Courtesy of D. I. Stuart, J. Grimes, P. Gouet, J. Diprose, R. Malby, P. Roy, B. P. V. Prasad and P. P. C. Mertens). |
Physicochemical and physical properties
The virion Mr is about 10.8×107, and the core Mr is about 6.7×107. Their buoyant densities in CsCl are 1.36 g cm−3 (virions) and 1.40 g cm−3 (cores). The S20,W is 550S (virions) and 470S (cores). Virus infectivity is stable at pH 8–9 but virions have a marked decrease in infectivity outside the pH range 6.5–10.2. In part, this may be related to the loss of outer coat proteins, particularly at the lower pH range. The sensitivity of the outer capsid proteins and their removal by cation treatment (e.g., by treatment with MgCl2, or CsCl) varies markedly with both pH and virus strain. At pH less than 5.0, virions and cores are both disrupted. Unlike orthoreoviruses, virus infectivity is abolished at pH 3.0. In blood samples, serum or albumin, virions held in vitro at less than 15 °C may remain infectious for decades. Purified BTV virions held at 4 °C in 0.1 M Tris/HCl pH 8.0 have no significant reduction in infectivity after 1 year. Crystals of core particles are very stable when kept at 29 °C. Virus infectivity is rapidly inactivated on heating to 60 °C. In general, orbiviruses are considered to be relatively resistant to treatment with solvents or detergents, although the sensitivity to specific detergents varies with virus species. However, sodium dodecyl sulfate will disrupt the particle and destroy its infectivity. Freezing reduces virus infectivity by about 90%, possibly due to particle disruption. However, once virus is frozen and held at −70 °C, infectivity remains stable.
Nucleic acid
The genomic dsRNA represents 12% or 19.5% of the total molecular mass of virions or cores, respectively. The genome is composed of 10 linear dsRNA segments that are packaged in equimolar ratios, one of each segment per virion. The genomic RNA is packaged as a series of ordered concentric shells within the VP3 layer of the subcore (Figure 2.Orbivirus). Four layers of RNA, each of which has elements of icosahedral symmetry, can be detected by X-ray crystallography of the BTV core. Within the central space of the subcore, there appears to be an association between the dsRNA molecules and the protein density at the five-fold axes of symmetry (at the vertices of the icosahedron), which is thought to represent the transcriptase complex (TC). From the five-fold axis, the RNA, in the outmost layer, appears to spiral away from the five-fold axes outward around the TC for two turns until it clashes with an icosahedrally related neighbor. At this point, it is thought to turn inward forming the next concentric shell of RNA. Genomic RNA contains 5′-terminal cap 1 structures (7mGpppG(2-Om)…).
For BTV, the genome segments range from 3,954 to 822 bp (total 19.2 kbp, total Mr 13.1×106). There is no evidence for short ssRNA oligonucleotides in intact virions. The genomic RNAs are numbered in order of decreasing Mr and increasing electrophoretic mobility in 1% agarose gels. For BTV, the segments migrate as three length classes: three long (Seg1–3: 3.9–2.8 kbp), three medium (Seg4–6: 2.0–1.6 kbp) and four short segments (Seg6–10: 1.2–0.8 kbp). For other members of the genus, different lengths and length classes exist. For viruses of an individual species, the dsRNA lengths from different isolates or serotypes are usually comparable, such that a uniform segment migration pattern is observed when the genomic RNAs of such isolates are analyzed by agarose gel electrophoresis. However, variations in primary sequence cause significant variations in rate and order of migration of genome segments during polyacrylamide gel electrophoresis (PAGE), particularly in high percentage gels (>5% polyacrylamide). Earlier BTV genome segment nomenclature based on PAGE is inconsistent, and the migration of Seg5 and Seg6 is often reversed.
With the exception of genome segments 9 and 10 which are bigenic, orbivirus genome segments have a single major ORF, which is always on the same strand (see conserved terminal sequences below). However, the main ORF may have more than one functional initiation site near to the 5′-end of the RNA, resulting in production of two related proteins.
For the isolate BTV-10, the 5′-non-coding regions (NCRs) of the positive-sense RNA strands range from 8 to 34 bp, and the 3′-NCRs are 31 to 116 bp. For other serotypes and viruses, the lengths may be different. However, in general the 5′-NCRs are shorter than the 3′-NCRs. Most orbivirus NCRs have the conserved complementary terminal sequences 5′-GU …. AC-3′ (Table 1.Orbivirus). The NCRs of BTV include terminal sequences of 6 bp that are usually identical for all 10 dsRNA segments (although some variation does occur) and that are conserved between different BTV isolates. Other orbiviruses have terminal sequences that are not always identical to those of BTV and may not be conserved in all 10 segments (Table 1.Orbivirus).
Fully functional and infectious virions have been recovered by the introduction of all 10 mRNAs into BSR cells.
Table 1.Orbivirus Conserved orbivirus terminal sequences (positive-sense strand)
|
Virus species |
Virus name |
5′-end |
3′-end |
|
African horse sickness |
African horse sickness virus (AHSV) |
5′-GUU(A/U)A(A/U) |
AC(A/U)UAC-3′ |
|
Bluetongue virus |
bluetongue virus (BTV) |
5′-GUUAAA |
(A/G)CUUAC-3′ |
|
Chobar Gorge virus |
Chobar Gorge virus (CGV) |
5′-GUUUA(A/U) |
(A/G)(G/C)(A/C)UAC-3′ |
|
Epizootic hemorrhagic disease virus |
epizootic hemorrhagic disease virus (EHDV) |
5′-GUUAAA |
(A/G)CUUAC-3′ |
|
Equine encephalosis virus |
equine encephalosis virus (EEV) |
5′-GUU(U/A) |
A (U/A/G)(A/U/C)GUUAC-3′ |
|
Eubenangee virus |
Eubenangee virus (EUBV) |
5′-GU(U/A)(A/U)AA |
(A/C)(U/A/C)UAC-3′ |
|
Great Island virus |
Great Island virus (GIV) |
5′-GUAAA(A/U)(A/U/C) |
(A/G)(A/U/C)(C/G)(C/A/G)AC-3′ |
|
Ieri virus* |
Ieri virus (IERIV) |
5′-GUU(U/A)AA |
(A/G/C)(G/A/C)NUAC-3′ |
|
Palyam virus |
Palyam virus (PALV) |
5′-GU(A/U)AAA |
(A/G)CUUAC-3′ |
|
Peruvian horse sickness virus |
Peruvian horse sickness virus (PHSV) |
5′-GUUAAAA |
(A/G)(C/G)(A/G)UAC-3′ |
|
St Croix River virus |
St Croix River virus (SCRV) |
5′-(A/G)UAAU(G/A/U) |
(G/A/U)(C/U)(C/A)UAC-3′ |
|
Umatilla virus |
Umatilla virus (UMAV) |
5′-GUUU(A/U)A |
A(G/A)GAUAC-3′ |
|
Wad Medani virus |
Wad Medani virus (WMV) |
5′-GU(A/U)(A/U)AA |
N(G/A/C)CUAC-3′ |
|
Yunnan orbivirus |
Yunnan orbivirus (YOUV) |
5′-GUUAAA(A/U) |
N(G/A/C)(A/G)UAC-3′ |
|
Warrego virus |
Warrego virus (WARV) |
5′-GUAAA(A/U) |
(A/C/U)C(U/A)UAC-3′ |
|
Wallal virus |
Wallal virus (WALV) |
5′-GUAUA(A/U) |
A(C/A)(A/G)C(U/A/C)UAC-3′ |
|
(not classified) |
Andasibe virus (ANDV)** |
5′-GUUAAA |
(A/U)CUUAC-3′ |
* Based on genome segments 3 to 10.
** Based on genome segments 2 to 10.
Proteins
There are seven orbivirus structural proteins (VP1 to VP7; Table 2.Orbivirus). These proteins constitute 88% and 80.5% of the dry weight of virions and cores, respectively. In BTV, the outer capsid consists of 180 copies of the 111 kDa sail-shaped VP2 protein arranged as trimeric triskelion structures, and 360 copies of an interdispersed and underlying VP5 protein (59 kDa), arranged as 120 trimers (Figure 1.Orbivirus and Figure 2.Orbivirus). The electrophoretic migration order and nomenclature of proteins may vary in members of other orbivirus species. Both VP2 and VP5 of BTV are attached to VP7. The surface of the core particle consists entirely of 780 copies of VP7, which are arranged with T=13 l symmetry, as a network of hexameric and pentameric rings (in a near-perfect example of quasi-equivalence; Figure 2.Orbivirus). The VP7 trimers of the core surface can bind dsRNA molecules, although the functional significance of this binding remains undetermined. Beneath the VP7 layer, the subcore capsid shell is composed of 120 copies of VP3 arranged with T=2 symmetry, displaying geometrical or pseudo quasi-equivalence (Figure 2.Orbivirus). The VP3 (T2) capsid shell encloses the 10 dsRNA segments (Figure 2.Orbivirus), as well as the three minor structural proteins. The latter include: the 150 kDa VP1(Pol), which is the RdRP; the 76 kDa VP4(Cap), which forms functional dimers and has both guanylyl-transferase and two methyltransferase (Mtr) activities [Mtr 1 (forming the 7-methyl guanosine of the cap structure) and Mtr 2 (forming the 2-O-methyl-guanosine, as the terminal nucleotide of the RNA chain)]; and the 36 kDa VP6/VP6a(Hel), which binds ssRNA or dsRNA and has both helicase and NTPase activities.
Table 2.Orbivirus Genome segments and protein products of bluetongue virus serotype 10
|
Genome segment |
bp |
Location of ORF |
Protein* |
Protein mass (kDa) |
Protein copy number/particle |
Function (location) |
|
Seg1 |
3954 |
12-3917 |
VP1(Pol) |
149.6 |
10 |
RdRP |
|
Seg2 |
2926 |
20-2887 |
VP2 |
111.1 |
180 |
Trimeric, highly variable cell attachment protein that forms the outer layer of the capsid. Reacts with neutralizing antibodies, is involved in virulence and determines virus serotype. Readily cleaved by proteases. |
|
Seg3 |
2770 |
18-2720 |
VP3(T2) |
103.3 |
120 |
Forms the innermost protein capsid shell subcore capsid layer. Through RNA binding activity and interactions with minor internal proteins controls overall size and organization of capsid structure, T=2 symmetry, |
|
Seg4 |
2011 |
9-1970 |
VP4(Cap) |
76.4 |
20 |
Dimer, Mtr 1 and Mtr 2, capping enzyme (guanylyl-transferase) |
|
Seg5 |
1769 |
35-1690 |
NS1(TuP) |
64.4 |
0 |
Forms characteristic tubules of unknown function in the cell cytoplasm. |
|
Seg6 |
1638 |
30-1607 |
VP5 |
59.2 |
360 |
Variable trimeric protein which which forms inner layer of the outer capsid and may be glycosylated., Together with VP2, determines virus serotype. |
|
Seg7 |
1156 |
18-1064 |
VP7(T13) |
38.5 |
780 |
Trimeric protein with T=13 symmetry that forms outer core surface andcan bind dsRNA. In some viruses (African horse sickness virus [AHSV]) forms flat hexagonal crystals, involved in cell entry and core particle infectivity in adults and cells of vector insects. Reacts with “core neutralizing” antibodies and is an immunodominant virus species-specific antigen. |
|
Seg8 |
1124 |
20-1090 |
NS2(ViP) |
41.0 |
0 |
Phosphorylated matrix protein of viral inclusion bodies with ssRNA binding activity. May be associated with outer capsid |
|
Seg9 |
1046 |
16-102 113-102
182-412 |
VP6(Hel)VP6a
NS4 |
35.8 31.9
9.5 |
60
0 |
VP6 binds to ssRNA and dsRNA, and has helicase and NTPase activities NS4 is a virulence factor that counters innate immune defences in mammalian cells (in particular interferon α/β). |
|
Seg10 |
822 |
20-706
59-706 108-204 |
NS3
NS3a NS5 |
25.6
24.0 7.5 |
0
0 0 |
Membrane glycoproteins, involved in cell exit. Variable in AHSV and involved in determination of its virulence Modulates cell transcription/translation |
The cryoEM structure of BTV-1RSA virions at 7-Å reveals structural information concerning the VP2 and VP5 outer coat proteins. The VP2 triskelion is composed of three tip domains branching from a central hub domain. The hub domain contains three putative sialic acid-binding pockets. Experimental data indicate that binding of the sugar-moiety is important for BTV infection. The VP5 membrane-penetration trimer, located between the VP2 trimers, has a central coiled-coil α-helical bundle, similar to the fusion proteins of many enveloped viruses. Weak interactions between the VP5 trimer and the VP2 trimer were detected in the cryoEM density map. Similar interactions were also detected with the underlying core surface layer of VP7 trimers. It has been suggested that the surface of VP5 could unfurl, like an umbrella, during penetration and shedding of the coat to release the transcriptionally-active core particle.
X-ray diffraction studies indicate that the minor structural proteins are attached as a TC to the inner surface of the subcore layer [VP3 (T=2)] at the five-fold symmetry axes (at the vertices of the icosahedron). However, because there is only a single TC at each position, they do not have full icosahedral symmetry and it has not yet been possible to determine their organization at the atomic level.
The VP7 protein of some viruses, such as AHSV, can also form flat hexagonal crystals/arrays, typically up to 5 μm in diameter, within the cytoplasm of the infected cell. These are composed of flat sheets of hexameric rings, which appear similar to the hexagonal rings of trimers seen in the core-surface layer.
There are five distinct non-structural viral proteins produced in cells infected with BTV or other orbiviruses. The 64 kDa NS1 (TuP) protein forms tubules that vary in length up to 4 μm. Although NS1 tubules have an unknown function, they are regarded as a characteristic feature of orbivirus replication. These tubules may have a ladder-like structure, as observed for BTV and epizootic hemorrhagic disease virus (EHDV) (68 and 52 nm in diameter, respectively), or they may be finer (23 nm in diameter) and have a reticular cross-weave pattern, as for AHSV.
The 41-kDa NS2 (ViP) protein can be phosphorylated and is an important component of the matrix of VIBs, which are the site of virus replication and assembly. VIBs also contain relatively large amounts of the virus core proteins. NS2 (ViP) consists of two domains joined by a hinge region and in its phosphorylated form assembles into large multimeric complexes. NS2 (ViP) has ssRNA-binding activity, suggesting that it plays an active role in replication. In conjunction with other virus proteins, it is believed to be involved in the recruitment of viral mRNA for encapsidation. The NS3/NS3a proteins are two small, non-structural membrane proteins (25 and 24 kDa) translated from different in-frame initiation sites on a single ORF and are involved in the release of virions from cells. This function may be essential for dissemination of progeny virus, particularly from insect vector cells, which can become persistently infected and do not show cytopathic effect or high levels of cell death. In the process of particle release, the NS3 proteins are also released from the cell. In addition to the NS3 ORF, genome segment 10 encodes NS5 from an overlapping out-of-frame ORF.
NS4 localises to both nucleoli and cytoplasm. In the cytoplasm NS4 of BTV has been shown to interact with lipid droplets. NS4 has been described as a virulence factor and counter-defence protein which dampens the innate immune response, particularly the interferon response, in mammalian cells. During late stages of infection, NS4 relocalises to the cell membrane.
Lipids
Orbivirus particles may be intimately associated with membraneous cell debris. Mature virions can acquire a membrane envelope by budding through the cell membrane during the process of cell exit, producing membrane-enveloped virus particles (MEVPs). However, this membrane is thought to be transient or unstable and the virions are usually considered to be non-enveloped.
Carbohydrates
The BTV VP5 protein may be glycosylated. NS3 and NS3a can become glycosylated, forming high molecular weight products.
Genome organization and replication
BTV genome segments are usually monogenic but Seg9 and Seg10 mRNAs both have two in-frame AUG codons, either of which can be used to initiate translation. Coding assignments are shown in Table 2.Orbivirus. The significance of the two forms of the Seg9 and Seg10 gene products (NS3, NS3a; VP6, VP6a) is not known. In some cases, other virus proteins form morphologically defined structures in infected cells (e.g., the flat hexagonal crystals/arrays formed of VP7 of AHSVs), but these are of unknown functional significance. Seg9 of Great Island virus (GIV) has two overlapping, out-of-phase ORFs, the longest of which codes for VP6 (Hel). The second, shorter ORF codes for a 22 kDa protein (identified as NS4), which has similarities to known dsRNA-binding proteins.
Virus adsorption involves components of the outer capsid, although cell entry may also involve VP7 (T13). VP2 (and possibly also VP5) is involved in determination of virulence. VP5 may be involved in penetration of the cell membrane (release from endosomes into the cytoplasm), and the expressed protein can induce cell fusion. The outer capsid layer is lost during the early stages of replication. The transcription frequency of mRNA from individual genome segments varies, with more copies produced from the shorter segments, such that approximately equivalent amounts of mRNA (by weight) are transcribed from each genome segment.
Some details of virus replication are unknown. VIBs are considered to be the sites of morphogenesis of transcriptionally active virus cores containing dsRNA. The smallest particles containing RNA, which are observed in VIBs, appear to represent progeny subcore particles. The outer core protein (VP7) is added within the VIB and the outer-capsid proteins at the periphery of the VIB.
Virions are transported within the cell by specific interaction with the cellular cytoskeleton and can be released from the cell prior to lysis through interaction with membrane-associated NS3 proteins (Figure 2.Reovirales). There is also some evidence of a specific association between NS1 tubules and intact virions in the cell cytoplasm. In most mammalian cells, replication of orbiviruses leads to shut-off of host protein synthesis and usually results in cell lysis and the release of virions. However, in persistently infected insect cells (or γδT cells), there is no evidence for shut-off of host protein synthesis, of extensive cell lysis or a cytopathic effect. In some viruses (such as AHSV), NS3 is involved in the determination of virulence in the mammalian host and, by controlling virus dissemination within the insect, may at least partially determine their ability to transmit the virus (vector competence). Virions can leave viable mammalian cells by two distinct mechanisms: extrusion (involving cell membrane damage) and budding. Budding results in particles that have a transient membrane envelope, which is lost shortly after leaving the cells. Continuous release of virions from infected cells and re-infection appear to be features of orbivirus replication.
Biology
The infectivity of purified (disaggregated) BTV particles is equivalent to a particle to infectivity ratio of approximately 1000:1 in both mammalian and insect cell systems. However, core particle infectivity varies from being 1000-fold less than that of intact virions (baby hamster kidney cells) to non-infectious (Chinese hamster ovary cells) in mammalian systems, depending on the cell line used. NS1 of BTV has also been reported to induce a cross-serotype and protective cell-mediated immune response in the mammalian host. In some insect cells (KC cells, derived from Culicoides sonorensis Wirth & Jones 1957 biting midges) and adult vector insects, core particles are only slightly less infectious than intact virions (particle to infectivity ratio of 1900:1). Treatment of virion with chymotrypsin or trypsin results in production of ISVPs, in which VP2 is cleaved. BTV ISVPs lack hemaglutinating activity, as well as the tendency to aggregate, but have a significantly elevated infectivity for adults of insect vectors and some insect cell lines (a particle to infectivity ratio of approximately 13:1 for KC cells).
Different orbiviruses infect a wide range of vertebrate hosts including domesticated and wild ruminants and equids, as well as rodents, bats, marsupials, birds, sloths and primates, including humans (Table 3.Orbivirus). Orbiviruses can also replicate in, and are primarily transmitted by, arthropod vectors (gnats, mosquitoes, phlebotomine sandflies or ticks, depending on the virus). Trans-stadial transmission in ticks has been demonstrated for some viruses. Infection of vertebrates in utero may also occur. Orbiviruses, particularly those transmitted by short-lived vectors (gnats, mosquitoes and phlebotomine sandflies), are only enzootic in areas where adults of the competent vector species persist and are present all, or most, of the year. Orbivirus RNA has been detected in Culicoides larvae recovered from outbreak areas, although trans-ovarial transmission has not been confirmed by the recovery of infectious virus. Orbiviruses have also been detected in cell lines derived from tick eggs. BTV and EHDV are distributed worldwide between about 50° north and 30° south in the Americas and between 40° north and 35° south in the rest of the world. These limits have recently expanded to 53° north (e.g., in Europe), possibly as the result of climate change.
There is also evidence for persistence of these viruses over winter in the absence of overt disease. Mechanisms for persistence in the vertebrate host species even at low levels (including vertical transmission in the vertebrate host) may be of particular importance. Virus distribution also depends on the initial introduction into areas containing susceptible vertebrate hosts and competent vector species. Not all serotypes of each virus (e.g., BTV) are present at locations where some serotypes are endemic. The relative abundance of different endemic strains can also vary significantly.
Orbivirus infection of arthropods has little or no overt effects. Infection in vertebrates can vary from inapparent to fatal, depending on both the virus and the host. Some BTV strains cause death in sheep; others cause a variety of pathologies, including hemorrhagic conditions, lameness, oedema, a transitory cyanotic appearance of the tongue (giving rise to the species name), nasal and mouth lesions, etc., or no overt pathology. BTV infected cattle may show no signs of disease but can involve long-lived viraemias. AHSV, EHDV and EEV can cause severe pathology in their respective vertebrate hosts. Mortality rates in serologically naive populations can be over 98% (AHSV). BTV is teratogenic and can cause severe deformaties (including dummy calf syndrome) due to a failure of the central nervous system to develop correctly after infection in utero in cattle and sheep.
Table 3.Orbivirus Orbivirus vectors and host range
|
Virus species |
Virus names (abbreviations) |
Vector (hosts) |
Serotypes |
|
African horse sickness virus |
African horse sickness virus (AHSV) |
Culicoides biting midges (equids, dogs, elephants, predatory carnivores and (in special circumstances) humans |
9 |
|
Bluetongue virus |
bluetongue virus (BTV) |
Culicoides biting midges (dogs, camels, cattle, sheep, goats, lynx) |
28 |
|
Changuinola virus |
Changuinola virus (CGLV), Almeirim virus, Altamira virus, Canindé virus, Gurupi virus, Irituia virus, Jamanxi virus, Jari virus, Monte Dourado virus, Ourém virus, Purus virus, Saracá virus |
Phlebotomine sandflies, culicine mosquitoes (humans, rodents, sloths) |
12 |
|
Chenuda virus |
Chenuda virus (CNUV), Baku virus, Essaouira virus, Huncho virus, Kala Iris virus, Mono Lake virus, Sixgun city virus |
Ticks (seabirds) |
7 |
|
Chobar Gorge virus |
Chobar Gorge virus (CGV), Fomédé virus |
Ticks (bats) |
2 |
|
Corriparta virus |
Corriparta virus (CORV), Acado virus, CS109 virus, V654 virus, V370 virus, Jacareacanga virus |
Culicine mosquitoes (humans, rodents) |
6 |
|
Epizootic hemorrhagic disease virus |
Epizootic hemorrhagic disease virus (EHDV-1), Ibaraki virus (EHDV-2) |
Culicoides biting midges (cattle, sheep, deer, camels, llamas, wild ruminants, marsupials) |
7* |
|
Equine encephalosis virus |
equine encephalosis virus (EEV) |
Culicoides biting midges (equids) |
7 |
|
Eubenangee virus |
Eubenangee virus (EUBV), Ngoupe virus, Pata virus, Tilligerry virus |
Culicoides biting midges, anopheline and culicine mosquitoes (hosts unknown) |
4 |
|
Great Island virus |
Great Island virus (GIV), Above Maiden virus, Arbroath virus, Bauline virus, Broadhaven virus, Cape Wrath virus, colony virus, colony B North virus, Elliðaey virus, Foula virus, Great Saltee Island virus, Grímsey virus, Inner Farne virus, Kemerovo virus, Kenai virus, Kharagysh virus, Lipovník virus, Lundy virus, Maiden virus, Mill Door virus, Mykines virus, North Clett virus, North End virus, Nugget virus, Okhotskiy virus, Poovoot virus, Røst Islands virus, St Abb’s Head virus, Shiant Islands virus, Thormódseyjarklettur virus, Tillamook virus, Tindhólmur virus, TribeÄ? virus, Vearrøy virus, Wexford virus, Yaquina Head virus |
Argas, Ornithodoros, Ixodes ticks (seabirds, rodents, humans) |
36 |
|
Ieri virus |
Ieri virus (IERIV), Gomoka virus, Arkonam virus |
Mosquitoes (birds) |
3 |
|
Lebombo virus |
Lebombo virus (LEBV) |
Culicine mosquitoes (humans, rodents) |
|
|
Orungo virus |
Orungo virus (ORUV) |
Culicine mosquitoes (humans, camels, cattle, goats, sheep, monkeys) |
4 |
|
Palyam virus |
Kasba virus/ChÅ«zan virus (KASV), Abadina virus, Bunyip Creek virus, CSIRO village virus, D’Aguilar virus, Gweru virus, Kindia virus, Marrakai virus, Marondera virus, Nyabira virus, Palyam virus, Petevo virus, Vellore virus |
Culicoides biting midges, culicine mosquitoes (cattle, sheep) |
13 |
|
Peruvian horse sickness virus |
Peruvian horse sickness virus (PHSV) |
Mosquitoes (horses) |
|
|
St Croix River virus |
St. Croix River virus (SCRV) |
Ticks (hosts unknown) |
|
|
Umatilla virus |
Umatilla virus (UMAV), Llano Seco virus, Minnal virus, Netivot virus |
Culicine mosquitoes (birds) |
4 |
|
Wad Medani virus |
Wad Medani virus (WMV), Seletar virus |
Boophilus, Rhipicephalus, Hyalomma and Argas ticks (domesticated animals) |
2 |
|
Wallal virus |
Wallal virus (WALV), Mudjinbarry virus, Wallal K virus |
Culicoides biting midges (marsupials) |
3* |
|
Warrego virus |
Warrego virus (WARV), Mitchell river virus, Warrego K virus |
Culicoides biting midges, anopheline and culicine mosquitoes (marsupials) |
3* |
|
Wongorr virus |
Wongorr virus (WGRV), Paroo River virus, Picola virus, MRM13443 virus, V199 virus, V595 virus, V1447 virus |
Culicoides biting midges, mosquitoes (cattle, macropods) |
8* |
|
Yunnan orbivirus |
únnán orbivirus (YUOV) |
Mosquitoes (cattle, sheep, donkeys) |
2 |
|
Unclassified |
Andasibe virus (ANDV) |
Mosquitoes (unknown) |
|
|
Unclassified |
Codajás virus (COV) |
Mosquitoes (rodents) |
|
|
Unclassified |
Guǎngxī orbivirus (GXOV) |
|
|
|
Unclassified |
Ife virus (IFEV) |
Mosquitoes (rodents, birds, ruminants) |
|
|
Unclassified |
Itupiranga virus (ITUV) |
Mosquitoes (unknown) |
|
|
Unclassified |
Japanaut virus (JAPV) |
Mosquitoes (unknown) |
|
|
Unclassified |
Kammavanpettai virus (KMPV) |
Unknown (birds) |
|
|
Unclassified |
Kemerovo virus (KEMV) |
|
|
|
Unclassified |
Lake Clarendon virus (LCV) |
Ticks (birds) |
|
|
Unclassified |
Matucaré virus (MATV) |
Ticks (unknown hosts) |
|
|
Unclassified |
mobuck virus (MBV) |
|
|
|
Unclassified |
Sathuvachari virus (SVIV) |
|
|
|
Unclassified |
Tembe virus (TMEV) |
Mosquitoes (unknown) |
|
|
Unclassified |
Tibet orbivirus (TIBOV) |
|
|
|
Unclassified |
Tracambe virus (TRV) |
Mosquitoes (unknown) |
|
*In some species the serological relationship between strains has not been fully determined.
Antigenicity
The main virus serogroup-specific antigen is the immunodominant outer core protein VP7. These virus serogroups correlate with virus species. Monoclonal or polyclonal antibodies against VP7 can neutralize core particle infectivity, but do not attach to, or neutralize, undamaged virions or infectious subvirion particles (ISVPs) in aqueous suspension, indicating that VP7 is not exposed on the intact virion surface. Other viral proteins are also conserved between members of a virus species, in particular the core proteins, NS1 and NS2. Some of these antigens may also show cross-reactivity with viruses in other species in the genus, particularly those regarded as closely related. These cross-reactions are usually at a significantly lower level than with other viruses from the same virus species and may be one-way. Such relationships between members of species are also demonstrated by comparisons of the RNA sequences of conserved segments; for example, homologs of BTV Seg3, coding for inner core protein VP3. These data indicate that orbiviruses may be divided into at least four groups that correspond to genomic groups. Group A contains: AHSV, BTV, EHDV, equine encephalosis virus (EEV), Eubenangee virus (EUBV), Palyam virus (PALV), Wallal virus (WALV) and Warrego virus (WARV). Group B contains: Chenuda virus (CNUV), Ieri virus (IERIV), Wad Medani virus (WMV) and GIV. Group C contains Corriparta virus (CORV). Group D contains Wongorr virus (WGRV). Insufficient comparisons have been made to conclusively assign all of the viruses of the genus Orbivirus to these groups.
For each orbivirus species, viruses can be divided into a number of serotypes that can be identified and distinguished in serum neutralization assays of intact virions, primarily via the specificity of interactions between neutralizing antibodies and the outer capsid proteins (CPs). VP2 is the main neutralization antigen of BTV. VP5 is also involved in determination of virus serotype, possibly by imposing conformational constraints on VP2. The VP2 and VP5 proteins of BTV show the greatest antigenic and sequence variation (Figure 3.Orbivirus). In other viruses (such as GIV), the relative sizes of the outer CPs (VP4 and VP5) are very different and their individual roles may also be different. There is evidence that VP2 of BTV and AHSV (particularly in association with VP5) and VP7 can act as protective antigens.
In AHSV, the small nonstructural proteins, NS3 and NS3a, are also variable and may be divided into three groups (α, β and γ) based on sequence analysis. Preliminary serological evidence suggests that NS3 cross-reacts poorly between these groups. NS3 can also be involved in the determination of virulence (for AHSV), possibly as a result of its involvement in the release of virions from cells (budding) and its consequent effect on virus dissemination. Recent sequencing studies of BTV NS3 also indicate that it can be highly variable, although this variation does not correlate with virus serotype.
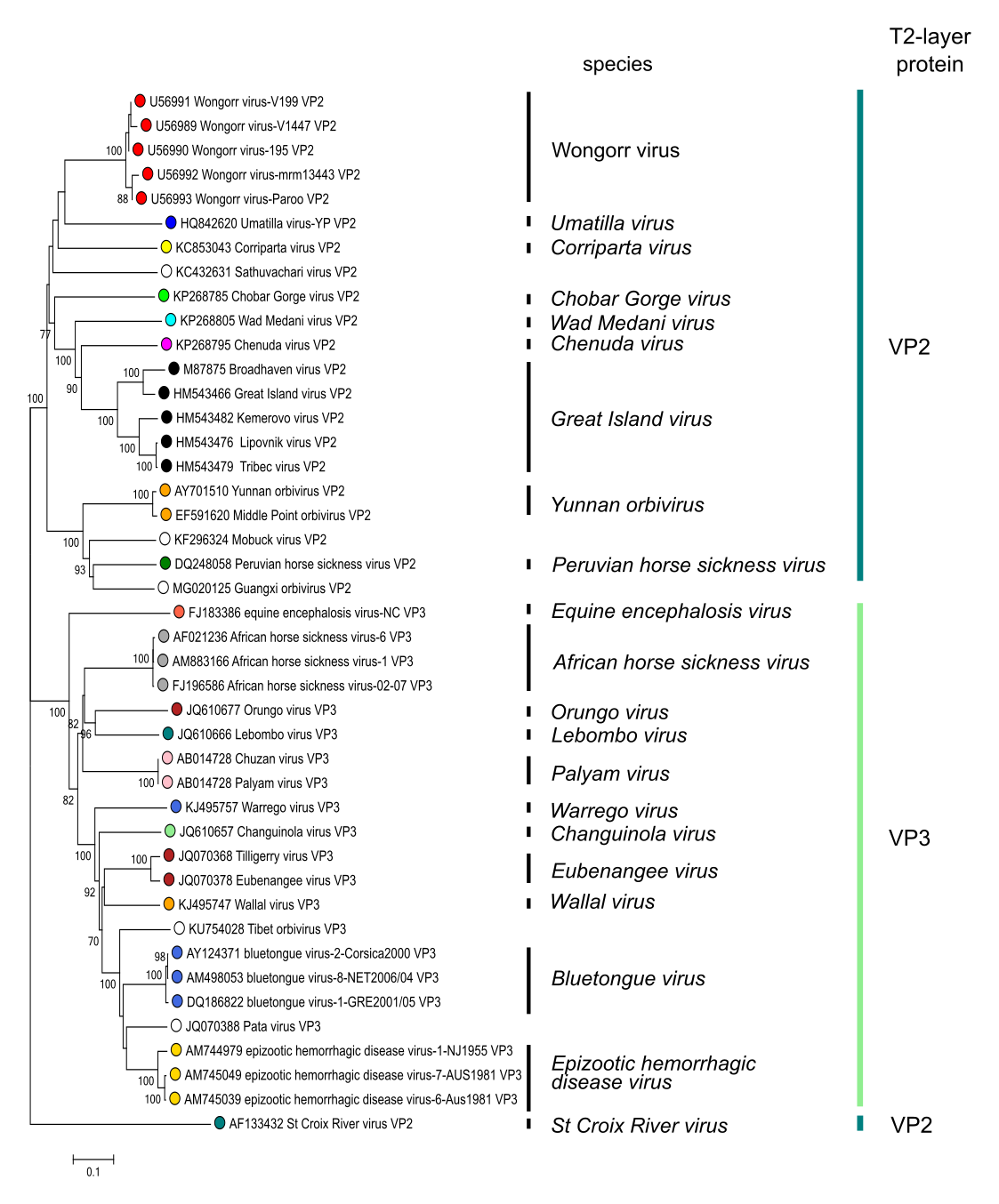 Phylogenetic tree of orbivirus T2 subcore shell proteins. The tree shows two groups: a mosquito-borne/tick-borne group where the second largest viral protein (VP2) forms the “T2” sub-core capsid layer; and a Culicoides miting midge-borne group where the third largest viral protein (VP3) forms the T2-layer. Sequences were aligned using MUSCLE (Edgar 2004) and the tree constructed in MEGA7 (Kumar et al., 2016) using the neighbour-joining method and the p-distance algorithm with pairwise deletion. Bootstrapping values (1000 replicates) above 70 % are shown. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. Phylogenetic tree of orbivirus T2 subcore shell proteins. The tree shows two groups: a mosquito-borne/tick-borne group where the second largest viral protein (VP2) forms the “T2” sub-core capsid layer; and a Culicoides miting midge-borne group where the third largest viral protein (VP3) forms the T2-layer. Sequences were aligned using MUSCLE (Edgar 2004) and the tree constructed in MEGA7 (Kumar et al., 2016) using the neighbour-joining method and the p-distance algorithm with pairwise deletion. Bootstrapping values (1000 replicates) above 70 % are shown. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Species demarcation criteria
In addition to the other general criteria used throughout the family, including compatability for reassortment, members of a species in the genus Orbivirus may be identified by:
- High levels of serological cross reaction using either polyclonal sera or monoclonal antibodies against conserved antigens such as VP7. For example, in competition ELISA at a test serum dilution of 1/5, a positive serum will show >50% inhibition of color formation, while a negative-control serum, or a serum that is specific for a different species, will normally produce <25% inhibition of color compared to a no-antibody control. Members of different species may show a low level serological cross-reaction, which may be only one-way.
- Sequence analysis: In the conserved Seg3 (encoding the major subcore structural protein, VP3), viruses within the same species will normally have >76% nucleotide identity (>83% amino acid identity) while those in different species usally have <74% identity. These differences are also reflected in the amino acid sequences of the viral proteins. In segment 1, encoding the RdRP, members of a single species will normally have >78% amino acid identity.
- Relatively efficient cross-hybridization of conserved genome segments (those not encoding outer capsid components or other variable proteins) under high stringency conditions (>85% identity).
- Common vector or host species and the pathology of infection. For example, BTV is transmitted only by certain Culicoides biting midges and will infect cattle and sheep, producing clinical signs of varying severity, but is not thought to infect horses. The reverse is true of AHSV in terms of mammalian host.
The phylogenetic relationships within the genus are illustrated in Figure 3.Orbivirus.
Related, unclassified viruses
|
Virus name |
Accession numbers |
Abbreviation |
|
Andasibe virus |
ANDV |
|
|
Codajás virus |
COV |
|
|
Guǎngxī orbivirus |
Seg1: MG020124; Seg2: MG020125; |
GXOV |
|
Ife virus |
IFEV |
|
|
Itupiranga virus |
ITUV |
|
|
Japanaut virus |
JAPV |
|
|
Kammavanpettai virus |
Seg1: MG770350; Seg2: MG770352; |
KMPV |
|
Kemerovo virus |
Seg1: HM543481; Seg2: HM543482; |
KEMV |
|
Lake Clarendon virus |
LCV |
|
|
Matucaré virus |
MATV |
|
|
mobuck virus |
Seg1: KF296322; Seg2: KF296323; |
MBV |
|
Sathuvachari virus |
Seg1: KC432629; Seg2: KC432631; |
SVIV |
|
Tembe virus |
TMEV |
|
|
Tibet orbivirus |
Seg1: KF746187; Seg2: KF746188; |
TIBOV |
|
Tracambe virus |
TRV |
Virus names and virus abbreviations are not official ICTV designations.
Based on phylogenetic analysis, Andasibe virus, Pata virus, Japanaut virus, Matucare virus, Guangxi orbivirus, Tibet orbivirus, Kemerovo virus, Sathuvachari virus, Mobuck virus and Kammavanpettai virus are likely to represent novel species.

