Family: Sedoreoviridae
Genus: Mimoreovirus
Distinguishing features
The mimoreovirus genome consists of 11 segments of linear dsRNA. The only virus assigned to the genus Mimoreovirus is Micromonas pusilla reovirus (MpRV), which was isolated from the marine Micromonas pusilla plant-like protist (algae) (Brussaard et al., 2004). MpRV does not grow in mammalian or fish cells lines but grows in the Micromonas pusilla LAC38 strain.
Virion
Morphology
Virions isolated from supernatants of infected Micromonas pusilla cells on Percoll® gradients have an average diameter of 90–95 nm (Figure 1.Mimoreovirus), larger than any previously described member of the order Reovirales. Some damaged particles have an outer layer of protein (about 15 nm thick) surrounding a more compact internal structure (about 75 nm in diameter).
MpRV virions purified on CsCl gradients have an average diameter of 75 nm, suggesting that they have lost the outer capsid proteins, although this size is similar to that of whole particles of other reoviruses. Twin bands of virions were recovered from CsCl gradients. The denser layer has traces of a protein with the same predicted mass as MpRV VP1. The protein content of these particles is similar to those of non-turreted intact particles such as rotaviruses, orbiviruses and seadornaviruses.
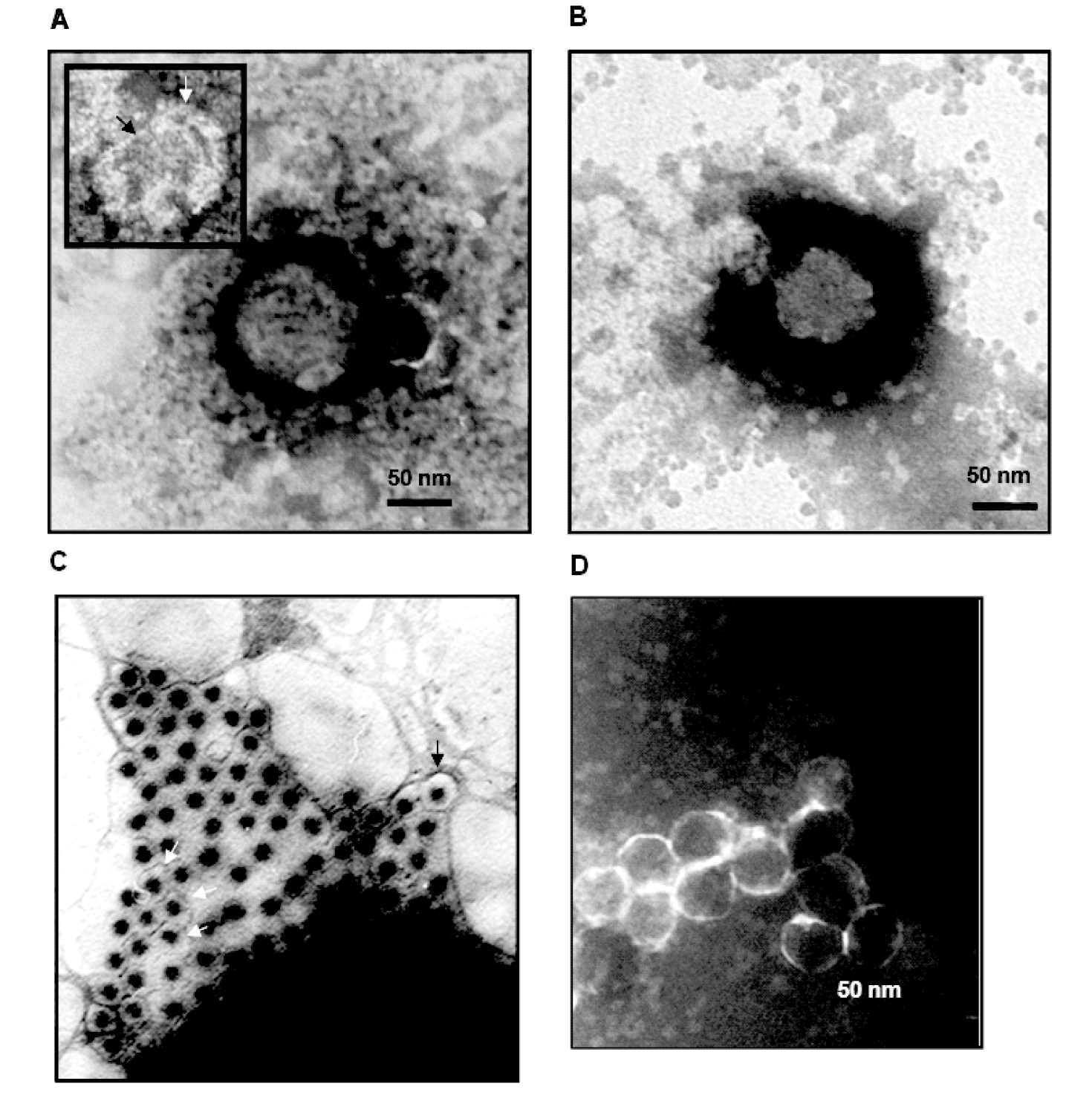 |
| Figure 1.Mimoreovirus (A) Micromonas pusilla reovirus virions purified on Percoll®, with a diameter of 95 nm. A damaged particle is shown at the upper left corner, showing an outer layer about 15 nm thick (indicated by a white arrow), surrounding a more compact structure with a diameter of about 75 nm (indicated by a black arrow). (B) A virion purified by CsCl gradient centrifugation, with a diameter of about 75 nm. (C) Particles pelleted from the clarified lyaste of infected Micromonas pusilla. Some particles (indicated by arrow) have a larger diameter. (D) Particles generated by treating whole virions (purified on Percoll®) with 1.5 M CaCl2 and subsequent purification on CsCl gradient, generating cores (or sub-cores) with smooth outline. (Courtesy of H. Attoui and C. Brussaard.) |
MpRV virions treated with CaCl2 and then purified on Percoll® gradients have a diameter of 50 nm, showing that they have lost outer capsid proteins and may have lost other components from the underlying capsid layer. These particles have a smooth outline (no turrets; Figure 1.Mimoreovirus), similar that observed for orbivirus (sub-core particles), rotaviruses and seadornaviruses.
The outer layer of the MpRV particle appears to represent a pseudo-envelope (or an additional coat) formed by the VP1 protein. Transient envelope structures have been described for orbiviruses, coltiviruses, rotaviruses and seadornaviruses as a consequence of the budding of virions from the cell membrane or budding into the endoplasmic reticulum during morphogenesis. MpRV may be the first member of the order Reovirales to possess a constitutive pseudo-envelope structure or an additional protein coat.
Physicochemical and physical properties
Thermostability of the virion has been tested at temperatures ranging from −196 °C (liquid nitrogen) to 95 °C. Virus is inactivated at temperatures above 35 °C. Freezing at temperatures below −20 °C preserved virion infectivity. Treatment with sodium dodecyl sulfate (0.1–0.5%) abolishes infectivity, while treatment with non-ionic detergents such as Tween 80, NP40 and triton X-100 (0.1–1%) does not affect infectivity. Exposure to acetone or alcohol inactivates the virion, while treatment with diethyl ether, chloroform or Vertrel XF preserves infectivity. These organic solvents might therefore be useful for virion purification. Exposure to acidic conditions (pH<5) inactivates the virus, while pH between 7 and 10 does not affect virus infectivity.
Virus replication is inhibited in Micromonas pusilla that are kept in the dark but resumes in the presence of light.
Nucleic acid
The genome consists of 11 dsRNA segments that are numbered in order of reducing Mr or increasing electrophoretic mobility during agarose gel electrophoresis (AGE). The genome of MpRV is 25,563 bp in total, with segment lengths ranging from 5,792 bp and 741 bp. Electrophoretic analysis (1% AGE) of genomic RNA shows a 1-1-2-5-2 migration pattern (Figure 2.Mimoreovirus).
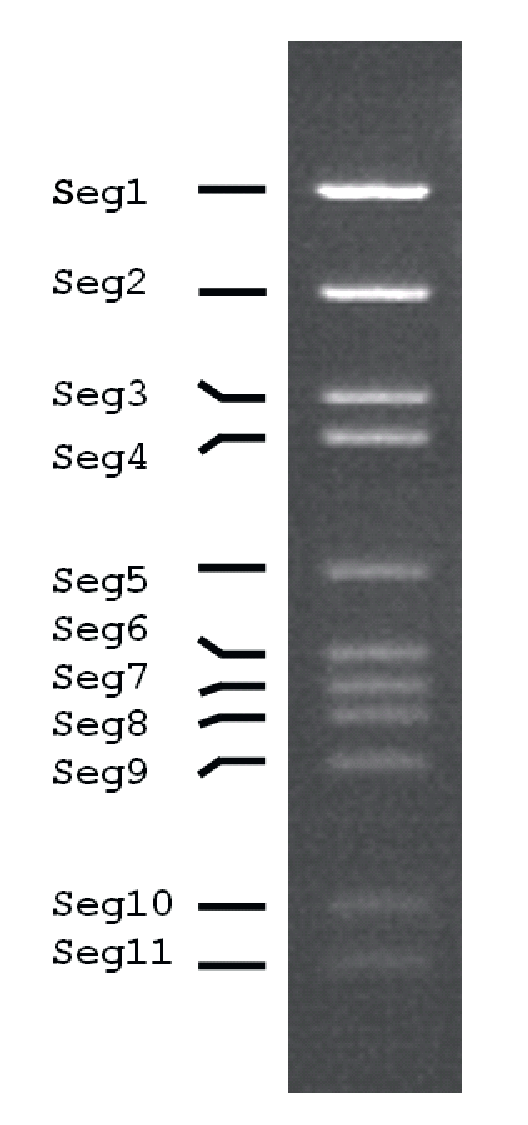 |
| Figure 2.Mimoreovirus Electrophoretic profile (electropherotype) of genome segments of Micromonas pusilla reovirus in 1% agarose gel. |
Sequence analysis of the MpRV genome (Attoui et al., 2006) has shown that each genome segment contains a single large ORF. The only exception is Seg5, which contains an ORF spanning nucleotide positions 44 to 2,005 that is interrupted by an in-frame “leaky” UGA stop codon at positions 1571–1573. The G+C content of MpRV genome segments is between 41 and 50%, the the highest value being observed for Seg5. The conserved terminal sequences of the 11 segments are: 5′-GAAGAA/U …. A/GAAAGUC-3′.
Proteins
Eight structural proteins are detected in virions purified by CsCl gradient centrifugation, their Mr being 200, 150, 120, 107, 67, 53, 35 and 32 kDa. The relative structural organisation of these proteins is not yet known and the role of the various structural and non-structural proteins is yet to be characterized although some putative assignments can be made based on sequence comparisons (Table 1.Mimoreovirus).
Carbohydrates
VP1 might be glycosylated based on it relatedness to mucin.
Genome organization and replication
Each of the 11 genome segments encodes a single protein except Seg5, where a “readthrough” inferred from sequence analysis may result in the production of two related proteins. The shorter 53.1 kDa protein, (VP5ter) would be produced by the early-termination of translation while the predicted readthrough protein (VP5rdt) would be 68.9 kDa. The other proteins encoded by the different genome segments are identified as VP1–VP4 and VP6–VP11 respectively. There are thus 12 predicted translation products (Table 1.Mimoreovirus).
Table 1.Mimoreovirus Genome segments and predicted protein products of Micromonas pusilla reovirus
|
Genome segment |
bp |
Protein |
Predicted protein mass (kDa) |
Structure/putative function |
|
Seg1 |
5792 |
VP1 |
201.35 |
Outer layer/pseudo-envelope |
|
Seg2 |
4175 |
VP2 |
154.69 |
RNA-directed RNA polymerase |
|
Seg3 |
3129 |
VP3 |
116.27 |
Sub-core “2” layer |
|
Seg4 |
2833 |
VP4 |
102.64 |
|
|
Seg5 |
2027 |
VP5(tr) / VP5(rdt) |
53.17 / 68.93 |
Has similarity to rotavirus VP4 outer capsid protein |
|
Seg6 |
1687 |
VP6 |
59.0 |
|
|
Seg7 |
1556 |
VP7 |
55.41 |
Has similarity to cypovirus NS1 |
|
Seg8 |
1449 |
VP8 |
51.69 |
Has similarity to rotavirus NSP2 |
|
Seg9 |
1296 |
VP9 |
44.31 |
Has similarity to nucleotide-binding core-protein of fijiviruses |
|
Seg10 |
878 |
VP10 |
24.69 |
|
|
Seg11 |
741 |
VP11 |
22.25 |
|
VP1 is considered likely to form the outermost surface of the virus, possibly representing an extra coat in comparison to other reoviruses. Between amino acid positions 88 and 255 VP1 is 24% identical with the minor capsid protein sigma-1 (a hemagglutinin and responsible for cell attachment) of mammalian orthoreovirus 3 (genus Orthoreovirus) and the sigma-c protein of Pulau reovirus (genus Orthoreovirus; 22% identityfor VP1 amino acid positions 172−321).
VP1 also has significant aa identity with viral and non-viral hemagglutinins, including those of the bacterial Burkholderia pathogens (amino acid identity 20%, similarity 40%) and Staphylococcus bacteria (identity 19%, similarity 38%), Candida albicans yeasts (identity 20%, similarity 39%) and baker’s yeast (Saccharomyces cerevisiae) (identity 20%, similarity 37%). VP1 also has similarities with proteins of some large DNA viruses such as: (i) phycodnavirids, including the surface glycoprotein Vp260 of Paramecium bursaria Chlorella virus (PBCV) (identity 24%, similarity 39%); (ii) herpesvirids, including the envelope protein glycoprotein 2 (gp2) of equine herpesvirus (identity 20%, similarity 32%); and (iii) bacteriophages, including the envelope protein of Acholeplasma phage L2 (family Plasmaviridae; identity 26%, similarity 46%). All of these glycoproteins are found in envelopes or cell wall structures.
MpRV VP1 has a high serine and threonine content (both ≥11%) compared to a value of 1 to 7.5 % for other amino acids in the protein. This is characteristic of glycoproteins and in particular, for mucin and mucin-like proteins and cell wall adhesins. Such serine- and threonine-rich proteins are usually heavily O-glycosylated. Amino acid sequence repeats within VP1 each align best (but not exactly) with a protein sequence immediately N-terminal to it suggesting that sequence duplication has been followed by separate evolution of the parental and daughter repeat sequences.
Seg2 of MpRV encodes VP2, which is thought to be the viral RNA-directed RNA polymerase. RdRP core motifs identified in the VP2 protein, include “SG” (amino acid positions 801–802) and “GDD” (amino acid positions 835–837). A partial match (amino acid positions 647–962, identity 21%) was also found within the enzyme “core” region, with the RdRP of human rotavirus C (accession number AJ304859), another 11 segmented dsRNA virus belonging to the order Reovirales.
Seg3 of MpRV encodes VP3, which by analogy with other reoviruses, appears likely to represent the structural protein which forms the inner capsid shell (sub-core). In other reoviruses this layer has been shown to have pseudo T=2 icosahedral symmetry (also described as a modified T=1 symmetry) and the sub-core shell protein is also identified as the T2 protein. VP3 was found to partially match the P3(T2) of the phytoreovirus rice dwarf virus (amino acid positions 229–311, identity 26%) and lambda-1(T2) of MRV3 (amino acid positions 50–145, identity 20%). The lambda-1 of MRV3 possesses NTPase and helicase activities.
Seg5 of MpRV encodes VP5, which at amino acid positions 214–318 is 21% identical to the outer capsid spike protein VP4 of rotavirus A (genus Rotavirus).
Seg7 of MpRV encodes VP7, which is 32% identical for amino acid positions 130–209 to the non-structural protein NS1 of Lymantria dispar cypovirus 1 (genus Cypovirus), while VP8 of MpRV (encoded by Seg8) is 28% identical for amino acid positions 42–66 to NSP2 of human rotavirus A, a protein with dsRNA helix destabilisation activity, that binds RNA and is an NTPase.
Seg9 of MpRV encodes VP9, which is 28% identical for amino acid positions 269–338 to the protein encoded by Seg7 of Nilaparvata lugens reovirus (genus Fijivirus), which is a core protein with nucleotide-binding activity.
There are no significant matches for VP4, VP6, VP10 and VP11.
Biology
MpRV was found in a sample of the marine Micromonas pusilla plant-like protist (algae), along with a larger virion (size 100–140 nm) with a dsDNA genome. However, the larger virion type was removed by passage through a 0.1 µm pore-size filter and end-point dilution. Ten virus clones all had comparable particle size, host specificity and infection parameters. The virus does not infect insect or mammalian cells and, out of six strains of M. pusilla, only strain LAC38 supports replication. LAC38 originates from Norwegian coastal waters, while the other five strains were isolated from different locations in the English Channel, English coastal waters, or the Gulf of Maine in the United States.
Growth of M. pusilla cells is inhibited within 24 hours by addition of MpRV The number of free virions starts to increase at 36 hours post infection, while a decline in algal cell numbers is observed by 40 hours post infection, the percentage of dead algal cells steadily increasing, to match an increase in virion released from the host cells (>60% dead cells). The burst size has been estimated at 460–520 virions per lysed algal cell.
Species demarcation criteria
There is only one species in the genus and species demarcation criteria have not been defined.
Related, unclassified viruses
|
Virus name |
Abbreviation |
|
|
Cimex lactularius reovirus |
ClRV |
|
|
Porcelio dilatatus reovirus |
||
|
Buthus occitanus reovirus |
Virus names and virus abbreviations are not official ICTV designations.
Cimex lactularius reovirus has 11 segments, an icosahedral double-layered capsid of about 50 nm in diameter and was isolated from the hemipteran bed bug Cimex lactularius Linnaeus, 1758. Porcelio dilatatus reovirus and Buthus occitanus reovirus are both uncharacterized and were isolated from a woodlouse (Porcellio dilatatus Brandt, 1833), and the common yellow scorpion (Buthus occitanus Amoreux, 1789), respectively (Eley et al., 1987).

