Family: Spinareoviridae
Genus: Cypovirus
Distinguishing features
In the cytoplasm of infected cells, cypovirus particles are enclosed individually or collectively within ‘polyhedra’ consisting of a crystalline matrix formed by a virus-encoded polyhedron protein. Cypoviruses only infect, and are pathogenic for, arthropods. Virions have single capsid shells with surface spikes, and have transcriptase and capping enzymes that are active without particle modification. They can retain RNA-directed RNA polymerase (RdRP) activity despite particle disruption into 10 distinct RNA protein complexes, each representing a single genome segment and a transcriptase complex. Consequently, transcriptase activity is resistant to repeated freeze–thawing, which disrupts the particle structure releasing the spikes from the particle surface. The transcriptase activity may have very pronounced dependence on the presence of S-adenosyl-L-methionine or related compounds, although this dependence may be reduced by repeated freeze–thawing and loss of spikes.
Virion
Morphology
Virions have a single-layered capsid, composed of a central capsid shell of 57 nm diameter, which extends to 71.5 nm (determined by cryoEM) when the 12 surface spikes or turrets that are situated on the icosahedral five-fold vertices are included. These surface projections are hollow and have been estimated to be up to 20 nm in length and 15–23 nm wide by conventional microscopy and negative-staining. These projecctions also appear to have a section near the tip that can be lost or removed. The virion has a central compartment about 35 nm in diameter. Cypovirions are structurally comparable to the core particles of members of other genera within the order Reovirales, particularly genera for viruses that produce particles with spiked cores (Orthoreovirus, Aquareovirus, Idnoreovirus and Oryzavirus) (Figure 1 Cypovirus and Figure 2 Cypovirus). The virions contain three major structural proteins that have been identified as: the capsid shell protein (CSP, 120 copies per particle), equivalent to VP3 (T2) of bluetongue virus and λ1 of orthoreoviruses; large protrusion protein (LPP, 120 copies per particle), comparable to orthoreovirus λ3 and turret protein (TP, 60 copies per particle), comparable to the orthoreovirus λ2. The virion also contains transcriptase enzyme complexes attached to the inner surface of the capsid shell at the icosahedral five-fold vertices.
Cypovirus particles may be occluded by a crystalline matrix of polyhedrin protein, forming a polyhedral inclusion body. These polyhedra have a symmetry (e.g., cubic, icosahedral or irregular) that is influenced by both the virus strain (polyhedrin sequence) and the host. The polyhedrin protein appears to be arranged as a face-centered cubic lattice with center-to-center spacing varying between 4.1 and 7.4 nm.
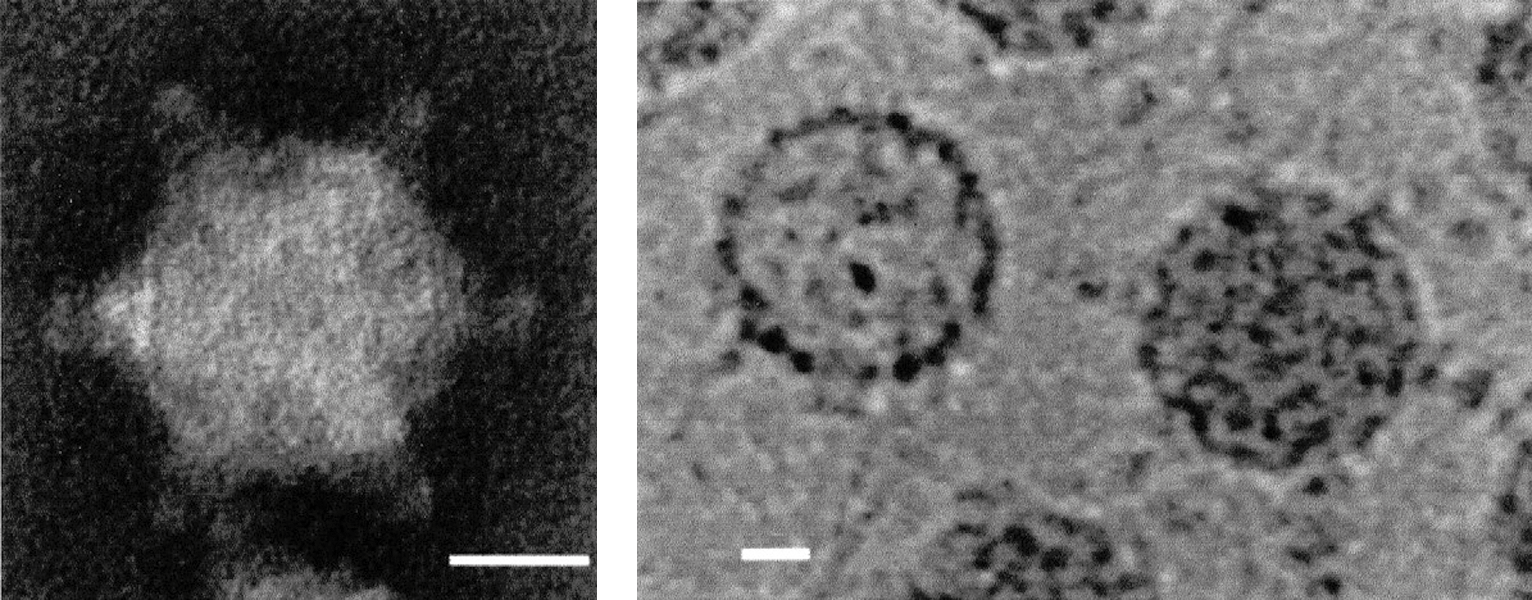 |
| Figure 1 Cypovirus (Left) Negative-contrast electron micrograph of a non-occluded virion of Orgyia psuedosugata cypovirus 5. (Right) Negative-contrast electron micrograph of empty and full “occluded” virions purified from polyhedra and stained with uranyl acetate (courtesy of C. L. Hill). The bars represent 20 nm. |
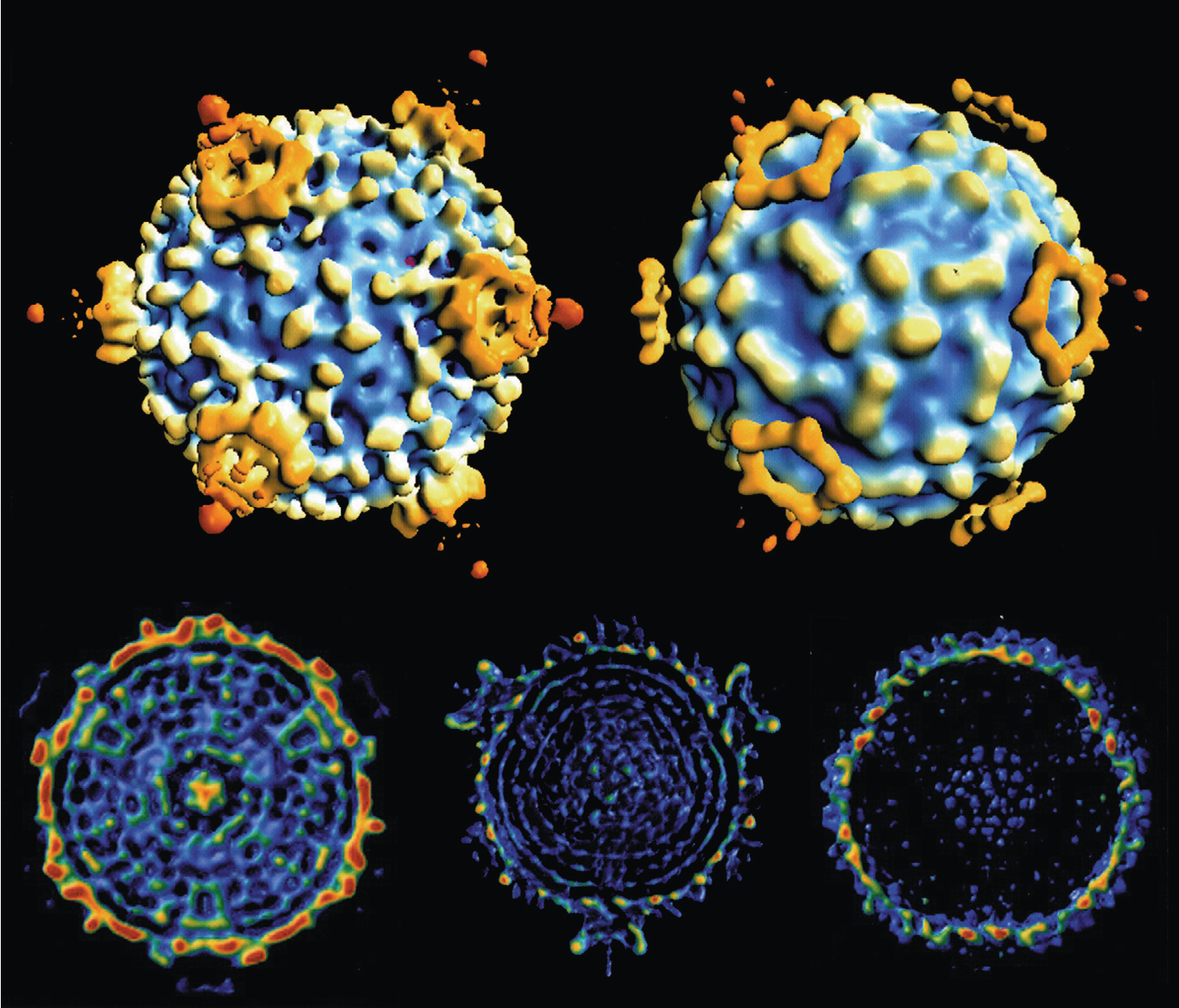 |
| Figure 2 Cypovirus CryoEM reconstructions of Orgyia psuedosugata cypovirus 5 virions, to 26 Å resolution: (top left) non-occluded virion; (top right) occluded virion; (bottom left) cross-section of a full occluded virion; (bottom center) cross-section of a full non-occluded virion; (bottom right) cross-section of an empty virion. The cross-sections show evidence of dsRNA packaged as distinct layers and suggest localization of the transcriptase complexes at the five-fold axes of symmetry (Courtesy of C. L. Hill). |
Physicochemical and physical properties
The virion Mr is about 5.4×107. The buoyant density in CsCl is 1.44 g cm−3 for virions, approximately 1.30 g cm−3 for empty particles, and 1.28 g cm−3 for polyhedra. The S20,W is approximately 420 S for virions and 260 S for empty particles. Polyhedra vary considerably in size and Mr and do not have a single characteristic S value. Polyhedra may occlude many virions or only single particles depending on the virus strain. Large polyhedra apparently containing no virions have also been observed.
Cypoviruses retain infectivity for several weeks at −15 °C, 5 °C or 25 °C. The virion retains full RdRP and capping activity after repeated freeze–thawing (up to 60 cycles). However, it appears that this may result in the loss of outer surface spikes or the breakdown of the virion into ten active and distinct enzyme/template complexes. Each complex contains one genome segment and a complete transcriptase complex, derived from the virion capsid (Yazaki and Miura 1980). Polymerase activity is therefore a poor indicator of virion integrity. Among members of the order Reovirales, the ability to retain enzyme function despite particle breakdown may be unique to cypoviruses.
Cations have relatively little effect on the virion structure, but incubation at 60 °C for 1 hour leads to degradation and release of genomic RNA. Virions are relatively resistant to treatment with trypsin, chymotrypsin, ribonuclease A, deoxyribonuclease or phospholipase. Virion enzyme functions also have some resistance to treatment with proteinase K. However, this may reflect the retention of enzyme activities despite particle disruption, particularly during the early stages of digestion. Cypovirus particles are resistant to detergent treatments such as 0.5–1% sodium deoxycholate but are disrupted by 0.5–1% SDS, which releases the genomic dsRNA. Treatment with Triton X-100, NP40 or urea also causes disruption of the virion structure. Fluorocarbon treatments have little effect on virion infectivity, while treatment with ethanol leads to the release of RNA. Virions and polyhedra are readily inactivated by UV-irradiation, which also releases the dsRNA template from individual genome segment/transcriptase complexes. Polyhedra remain infectious for years at temperatures below 20 °C. Virions can be released from polyhedra by treatment with carbonate buffer at pH >10.5 but are disrupted at pH <5. High pH treatment completely dissolves the polyhedral protein matrix, as in the mid-guts of permissive insects. This process is partly due to increased solubility of polyhedrin at high pH but is also aided by alkaline-activated proteases associated with polyhedra.
Nucleic acid
Polyhedra (but not virions) contain significant amounts of adenylate-rich oligonucleotides. In the majority of cases, cypovirus particles contain ten linear dsRNA genome segments. However, there is evidence to indicate that in some cases the virions may also contain an eleventh short segment (e.g., Trichoplusia ni cypovirus 15 [TnCPV15]). In Bombyx mori cypovirus 1 (BmCPV1), the genome segments vary from 4,190 to 944 bp with a total genome of 24,809 bp. Estimated genome segment lengths for other cypoviruses by electrophoretic comparisons are 0.6 to 5.6 kbp (Mr 0.42×106 to 3.7×106) with total genome lengths of 29.2 to 33.3 kbp (Mr 19.3×106 to 22.0×106).
The length distribution of genome segments (Table 1 Cypovirus) varies widely between different cypoviruses (e.g., the shortest dsRNA is estimated to vary between 530 and 1,440 bp). These length differences have formed a basis for the recognition and classification of distinct species of cypoviruses through the analysis of patterns of dsRNA migration (electropherotypes) during electrophoresis using 1% agarose or 3% SDS-PAGE which differ significantly in the migration of at least three genome segments, and frequently in the majority of segments. These patterns for members of the species Cypovirus altineae, Cypovirus autographae and Cypovirus lymantriae have some overall similarity, although in each case at least three segments show significant migrational differences. These viruses also show significant serological cross-reactions. More recently, it has been shown that members of different cypovirus species can also be distinguished on the basis of RNA sequence comparisons (e.g., by comparison of genome segment 10: the polyhedrin gene).
Table 1 Cypovirus Cypovirus genome segment lengths (kbp)
| Virus name | Total genome | Genome segment number | ||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
| Bombyx mori cypovirus 1 | 24.8 | 4.19 | 3.86 | 3.85 | 3.26 | 2.85 | 1.8 | 1.5 | 1.33 | 1.19 | 0.99 | |
| Inachis io cypovirus 2 | 25.5 | 4.06 | 4.06 | 3.83 | 3.65 | 2.21 | 1.93 | 1.79 | 1.56 | 1.38 | 0.98 | |
| Anaitis plagiata cypovirus 3 | 26.7 | 4.29 | 4.12 | 4.12 | 3.69 | 3.6 | 2.29 | 2.15 | 1.08 | 0.83 | 0.6 | |
| Antheraea assamensis cypovirus 4 | 25.0 | 3.85 | 3.79 | 3.78 | 3.35 | 2.18 | 1.94 | 1.79 | 1.68 | 1.48 | 1.14 | |
| Orgyia pseudotsugata cypovirus 5 | 26.3 | 4.17 | 4.17 | 4.17 | 3.69 | 3.22 | 2.17 | 2.06 | 1.21 | 0.88 | 0.88 | |
| Aglais urticae cypovirus 6 | 27.2 | 4.17 | 4.06 | 4 | 3.72 | 2.73 | 2.36 | 2.23 | 1.63 | 1.4 | 0.9 | |
| Mamestra brassica cypovirus 7 | 25.6 | 4.32 | 4.15 | 4.02 | 3.81 | 2.54 | 2.27 | 2.02 | 1.08 | 0.85 | 0.53 | |
| Abraxas grossulariata cypovirus 8 | 27 | 4.54 | 4.54 | 4.4 | 3.92 | 3.69 | 1.9 | 1.3 | 1.19 | 0.88 | 0.65 | |
| Agrotis segetum cypovirus 9 | 24.1 | 4.32 | 4.18 | 4.07 | 3.62 | 2.34 | 1.72 | 1.72 | 0.78 | 0,69 | 0.69 | |
| Aporophyla lutulenta cypovirus 10 | 27.6 | 4.31 | 4.31 | 4.02 | 4.02 | 2.5 | 2.29 | 2.29 | 1.69 | 1.21 | 0.99 | |
| Heliothis armigera cypovirus 11 | 25.5 | 4.6 | 4.4 | 4.4 | 3.83 | 1.98 | 1.98 | 1.35 | 1.27 | 0.98 | 0.71 | |
| Autographa gamma cypovirus 12 | 26.1 | 4.43 | 4.12 | 4.12 | 3.67 | 3.3 | 2 | 1.44 | 1.27 | 1.13 | 0.64 | |
| Polistes hebraeus cypovirus 13 | 25.2 | 4.26 | 4.26 | 4.03 | 3.6 | 3.2 | 1.6 | 1.4 | 1.14 | 0.98 | 0.78 | |
| Lymantria dispar cypovirus 14 | 25.3 | 4.33 | 4.06 | 3.92 | 3.34 | 3.16 | 1.78 | 1.39 | 1.25 | 1.14 | 0.96 | |
| Trichoplusia ni cypovirus 15 | 24.9 | 4.36 | 4.19 | 3.88 | 3.31 | 2.26 | 1.86 | 1.78 | 1.23 | 1.16 | 0.9 | 0.20* |
| Choristoneura occidentalis cypovirus 16 | 3.768 | 3.69 | 3.29 | 2.214 | 1.946 | 1.702 | 1.235 | 1.171 | ||||
| Uranotaenia sapphirina cypovirus 17** | 24.6 | 3.87 | 3.75 | 3.58 | 3.3 | 2.4 | 1.9 | 1.85 | 1.5 | 1.5 | 0.8 | |
| Operophtera brumata cypovirus 18** | 24.7 | 4.17 | 3.79 | 3.79 | 3.25 | 2.88 | 1.8 | 1.47 | 1.42 | 1.18 | 0.93 | |
| Operophtera brumata cypovirus 19** | 23.9 | 4.17 | 3.76 | 3.64 | 3.27 | 2.11 | 1.89 | 1.7 | 1.28 | 1.18 | 0.87 | |
| Simulium ubiquitum cypovirus 20** | 22 | 3.7 | 3.65 | 3.6 | 3.1 | 2.2 | 1.75 | 1.4 | 1.4 | 1.25 | 0.85 | |
Bold: length deduced from sequence analysis of the genome segment; plain font length estimated from electrophoretic comparsions. Previously published estimates of genome segment lengths for members of the species Cypovirus inachidis, Cypovirus aplocerae, Cypovirus antheraeae, Cypovirus betineae, Cypovirus aglaidis, Cypovirus mamestrae, Cypovirus gatineae, Cypovirus agrotidis, Cypovirus aprophylae, Cypovirus heliothidis, Cypovirus autographae and Cypovirus polistis have been adjusted in line with base pair values derived from sequencing studies of cDNA copies of genome segments from Bombyx mori cypovirus 1.
* Trichoplusia ni cypovirus 15 has been reported to contain an 11th short genome segment (200 bp).
** Unclassified isolates (see Table 2 Cypovirus).
The termini of the positive-sense strands are identical or very closely related for each of the different genome segments within members of the species Cypovirus altineae, but differ from those reported for members of other Cypovirus species (Table 2 Cypovirus). Choristoneura fumiferana cypovirus 16, (CfCPV16) has high levels of overall sequence variation compared to members of species Cypovirus altineae, Cypovirus inachidis, Cypovirus betineae, Cypovirus lymantriae or Cypovirus trichoplusiae and is therefore considered to be a member of a different species (Cypovirus choristoneurae), although its single sequenced segment (Seg10) has a similar 5′- and different 3′-end to representatives of species Cypovirus betineae. These data that different cypovirus electropherotypes usually have different conserved RNA terminal sequences.
Table 2 Cypovirus Conserved terminal sequences (positive-sense strand) of cypovirus genome segments
| Virus species | Virus name | 5′-end | 3′-end |
|---|---|---|---|
| Cypovirus altineae | Bombyx mori cypovirus 1 (BmCPV1) | 5′-AGUAA | GUUAGCC-3′ |
| Cypovirus altineae | Dendrolimus punctatus cypovirus 1 (DpCPV1) | 5′-AGUAA | GUUAGCC-3′ |
| Cypovirus altineae | Lymantria dispar cypovirus 1 (LdCPV1) | 5′-AGUA/GA/G | GU/CUAGCC-3′ |
| Cypovirus inachidis | Inachis io cypovirus 2 (IiCPV2) | 5′-AGUUUUA | UAGGUC-3′ |
| Cypovirus antheraeae | Antheraea assamensis cypovirus 4 (AaCPV4) | 5′-AGUAAU | AUAGAGC-3′ |
| Cypovirus antheraeae | Antheraea mylitta cypovirus 4 (AmCPV4) | 5′-AGUAAUCGACG | UAGAGC-3' |
| Cypovirus betineae | Orgyia pseudotsugata cypovirus 5 (OpCPV5) | 5′-AGUUU/AU/A
| UUGC-3′ |
| Cypovirus lymantriae | Lymantria dispar cypovirus 14 (LdCPV14) | 5′-AGAA | CAGCU-3′ |
| Cypovirus trichoplusiae | Trichoplusia ni cypovirus 15 (TnCPV15) | 5′-AUUAAAAA | GC-3′ |
| Cypovirus choristoneurae | Choristoneura occidentalis cypovirus 16 (CoCPV16) | 5′-AGUUUA/U | AAA/UUUUGUGC-3′ |
| unclassified | Uranotaenia sapphirina cypovirus 17 (UsCPV17) | 5′-AGAACAAA | UACACU-3′ |
| unclassified | Operophtera brumata cypovirus 18 (ObCPV18) | 5′-AGUAAAG/U/AC/U | U/CA/CGUUAGCU-3′ |
| unclassified | Operophtera brumata cypovirus 19 (ObCPV19) | 5′-AACAAAA/UA/U | A/UGA/UUUUGC-3′ |
| unclassified | Simulium ubiquitum cypovirus 20 (SuCPV20) | 5′-AGAAAAC | CAUGGC-3′ |
| unclassified | Maruca vitrata cypovirus (MvCPV21) | 5′-AUAUAAUU | AGUUAGU-3′ |
Proteins
Cypovirus particles generally contain five to six distinct proteins, two or three with a mass of more than 100 kDa. For BmCPV1, the structural proteins are VP1 (148 kDa), VP2 (136 kDa), VP3), VP4 (120 kDa), VP6 (64 kDa) and VP7 (31 kDa). Polyhedra also contain a 25–37 kDa polyhedrin protein (28.5 kDa for BmCPV1) that constitutes about 95% of the polyhedra protein dry weight. Due to the very high level of variation between different cypoviruses, it is unlikely that their homologous proteins will be identifiable simply by their migration order during PAGE.
Lipids
Cypoviruses are not known to contain any lipids in either virions or polyhedra.
Carbohydrates
The polyhedrin protein is glycosylated.
Genome organization and replication
Coding assignments for the genome segments of BmCPV1 are indicated in Table 3 Cypovirus. The cognate genes of other cypoviruses are not known. The large variations in the lengths of genome segments between most cypoviruses (apart from members of the species Cypovirus altineae, Cypovirus autographae and Cypovirus lymantriae) indicate relatively distant relationships and suggest that these assignments will not apply to members of other cypovirus species. Genome segment coding assignments generated by in vitro translation of individual denatured genome segment RNAs have been published for members of Cypovirus altineae and Cypovirus inachidis. These data and subsequent sequencing studies indicate that, in many cases, polyhedrin may be encoded by the shortest segment.
Table 3 Cypovirus Genome segments and protein products of Bombyx mori cypovirus 1
| Genome segment | bp | Protein1 nomenclature (2) | Predicted mass (kDa) | Function (location) |
|---|---|---|---|---|
| Seg1 | 4190 | VP1 (VP1) | 148 | Major capsid protein (CP) (virion) |
| Seg2 | 3854 | VP2 (VP2) | 136 | RdRP (virion) |
| Seg3 | 3846 | (VP3) | 140 | (virion) |
| Seg4 | 3262 | VP3 (VP4) | 120 | Possible methyltransferase and guanylyltransferase (virion) |
| Seg5 | 2852 | NS1 (NS5) | 101 | Non-structural, contains auto cleavage aa sequence, similar to foot-and-mouth disease virus 2Apro |
| NS2 (NS5a) | 80* | |||
| NS6 (NS5b) | 23* | |||
| Seg6 | 1796 | VP4 (VP6) | 64 | Leucine zipper ATP/GTP binding protein (virion) |
| Seg7 | 1501 | NS3 | 50 (61*) | Non-structural, with “structural” cleavage products |
| NS4 | 58* | |||
| (VP7) | ||||
| Seg8 | 1328 | VP5 or P44 (NSP8) | 44 | Unknown (anomalous migration during PAGE, with apparent mass 55 kDa) |
| Seg9 | 1186 | NS5 (NSP9) | 36 | Non-structural, dsRNA binding |
| Seg10 | 944 | Polyhedrin (Pod) | 28.5 | Polyhedron matrix protein (Pod) |
Length of genome segments and mass of encoded proteins deduced from sequence analysis
* Masses of proteins estimated from electrophoretic migration.
1 Protein nomenclature suggested by McCrae and Mertens (McCrae and Mertens 1983).
2 Alternative nomenclature suggested by (Hagiwara et al., 2002).
Unlike orthoreoviruses, cell entry and initiation of cypovirus replication in insect cells does not require modification of virions for activation of core-associated transcriptase enzymes. Uptake appears to be a relatively inefficient process in cell culture, which can be very significantly improved by the use of liposomes. Virus replication and assembly occur in the host cell cytoplasm, although there is some evidence that viral RNA synthesis may also occur in the nucleus. Replication is accompanied by the formation of viroplasms (viral inclusion bodies or virogenic stroma) within the cytoplasm. Viroplasms contain large amounts of virus proteins and virions. How genome segments are selected for packaging and assembly into progeny particles is not known. The importance of the terminal regions in this process is indicated by the packaging and transcription of a mutant Seg10 from an isolate of Bombyx mori cypovirus 1 (species Cypovirus altineae) that contained only 121 bp from the 5′-end and 200 bp from the 3′-end (Arella et al., 1988). Particles are occluded within polyhedra, apparently at the periphery of the virogenic stroma, from about 15 hours post infection onwards. The polyhedrin protein is produced late in infection and in large excess compared to other viral proteins. It is not known how polyhedrin synthesis is regulated.
Biology
Cypoviruses have only been isolated from arthropods. Attempts to infect vertebrate or vertebrate cell lines have failed. In addition, cypovirus replication is inhibited at 35 °C. Even susceptible insect larvae incubated with cypoviruses fail to develop infections at temperatures ≥35 °C. Cypoviruses are normally transmitted by ingestion of polyhedra on contaminated food materials. The polyhedra dissolve within the high pH environment of the insect gut and release occluded virions which then infect the cells lining the gut wall. Virus infection in larvae is generally restricted to the columnar epithelial cells of the midgut, although goblet cells may also become infected. Cypovirus replication in the fat body has been reported. In larvae, the virus infection spreads throughout the midgut region. In some species the entire gut is occasionally infected. The production of very large numbers of polyhedra gives the gut a characteristically creamy-white appearance. In infected cells, the endoplasmic reticulum is progressively degraded, mitochondria enlarge and the cytoplasm becomes highly vacuolated. In most cases, the nucleus shows few pathological changes. An exception is one little-studied Bombyx mori cypovirus strain that produces inclusion bodies within the nucleus (Kawase and Yamaguchi 1974). In the later stages of infection, cellular hypertrophy is common and microvillae are reduced or completely absent. Very large numbers of polyhedra are released by cell lysis into the gut lumen and excreted. The gut pH is lowered during infection and this prevents dissolution of progeny polyhedra in the gut fluid.
The majority of cypovirus infections produce chronic disease, often without extensive larval mortality. Consequently, many individuals reach the adult stage even though heavily diseased. However, cypovirus infections produce signs of starvation due to changes in the gut cell structure and reduced adsorptive capacity. Infected larvae stop feeding as early as 2 days post infection. Larval body size and weight are often reduced and diarrhea is common. The larval stage of the host can be prolonged by a factor of about 1.5. The size of infected pupae is frequently reduced, and the majority of diseased adults are malformed. They may not emerge correctly, and may be flightless. Infected females may exhibit a reduced egg-laying capacity.
Virus can be transmitted on the surface of eggs, producing high levels of infection in the subsequent generation. However, provided the egg surface is disinfected, no transovarial transmission has been observed. The infectious dose increases dramatically in the later larval instars. Different virus strains vary significantly in virulence. Larvae can recover from cypovirus infection, possibly because the gut epithelium has considerable regenerative capacity and because infected cells are shed at each larval molt.
Antigenicity
Serological cross-comparisons of cypovirus structural and polyhedrin proteins support the use of genomic dsRNA electropherotypes as one of the species demarcation criteria for the genus Cypovirus. Virus isolates within a single electropherotype have high levels of antigenic cross-reaction (in both polyhedrin and virion structural proteins), and efficient cross-hybridization of denatured genomic RNA, even under high-stringency conditions. In contrast, there is evidence of little or no serological cross-reaction between viruses representing different electropherotypes. Exceptions are members of the species Cypovirus altineae and Cypovirus autographae, which show low-level serological cross-reactions but also have some overall similarity in electropherotype patterns and show a low level of cross-hybridization of their genome segments. Member of the species Cypovirus lymantriae also have some similarity in RNA electropherotype patterns to viruses in both Cypovirus altineae and Cypovirus autographae and may therefore also have some antigenic relationship and RNA sequence similarity with these viruses.
Species demarcation criteria
Cypoviruses are classified into 16 species, most of which were initially established based on the distinctive dsRNA electropherotype patterns of their members. Cross-hybridization analyses of the dsRNA, serological comparisons of cypovirus proteins and, more recently, comparison of RNA sequences (Figure 3 Cypovirus) have confirmed the validity of this classification, but have identified the need for additional virus species.
 |
| Figure 3 Cypovirus Phylogenetic relationships between cypovirus isolates using the amino acid sequences of the polyhedrin protein. Sequences were aligned using MUSCLE (Edgar 2004) and the tree constructed in MEGA7 (Kumar et al., 2016) using the neighbour-joining method and the p-distance algorithm with pairwise deletion. Bootstrapping values (1000 replicates) above 70 % are shown. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
For example, different isolates of Orgyia pseudotsugata cypovirus 5 (species Cypovirus betineae) are >98% identical in genome Seg10 (the polyhedrin gene), while isolates of Bombyx mori cypovirus 1, Dendrolimus punctatus cypovirus 1 (DpCPV1) and Lymantria dispar cypovirus 1 (species Cypovirus altineae) have 89–98% nucleotide sequence identity in this gene. In contrast, polyhedrin sequence comparisons of members of unrelated species showed only low levels of sequence identity (20–23%). Studies of genomic RNA from different cypovirus species have demonstrated that, although there may be slightly higher conservation in the longest genome segments (possibly as a result of functional constraints), the level of variation is relatively uniform across the whole genome. This contrasts with reoviruses that infect vertebrates, perhaps because there is no neutralizing antibody response in the host insects of cypoviruses and consequently no selective pressure to select for variation in outer capsid proteins.
In addition to the other general criteria used throughout the family, members of a species in the genus Cypovirus may be identified by:
- Similar electrophoretic migration of at least seven genome segments, as analyzed using either an agarose, or a low percentage (3%) polyacrylamide gel system. Viruses of different species have significant migrational differences in at least three genome segments.
- Nucleotide sequence conservation >80% in genome segment 10 for instance.
- Cross-hybridization of genome segments under high stringency conditions designed to detect >90% identity.
Virus names include both the host species from which the virus was first isolated, and a numerical suffix derived from the species name. For example, Bombyx mori cypovirus 1 (BmCPV1) was first isolated from the silkworm Bombyx mori (Linnaeus, 1758) and is a member of the species Cypovirus altineae.
Related, unclassified viruses
| Virus name | Accession numbers | Abbreviation |
| Uranotaenia sapphirina cypovirus 17 | Seg10: AY876384 | UsCPV17 |
| Culex restuans cypovirus 17 | Seg10: DQ212785 | CrCPV17 |
| Operophtera brumata cypovirus 18 | Seg5: DQ192245; Seg6: DQ192246; Seg7: DQ192247; Seg8: DQ192248; Seg9: DQ192249; Seg10: DQ192250 | ObCPV18 |
| Operophtera brumata cypovirus 19 | Seg2: DQ192251; Seg5: DQ192252; Seg9: DQ192253; Seg10: DQ192254 | ObCPV19 |
| Simulium ubiquitum cypovirus 20 | Seg10: DQ834386 | SuCPV20 |
| Maruca vitrata cypovirus 21 | MvCPV21 | |
| Heliothis armigera cypovirus (“B” strain) | HaCPV-B | |
| Maruca vitrata cypovirus (A strain) | MvCPV-A | |
| Maruca vitrata cypovirus (B strain) | MvCPV-B | |
| Plutella xylostella cypovirus | PxCPV |
Virus names and virus abbreviations, are not official ICTV designations.
More than 230 cypovirus-related viruses have been described from hosts such as lepidoptera and hymenoptera, but also one from a freshwater daphnid. Given the known diversity of insect species, there are likely to be a very large number of cypovirus species.

