Family: Spinareoviridae
Genus: Dinovernavirus
Distinguishing features
The dinovernavirus genome consists of nine segments of dsRNA. Aedes pseudoscutellaris reovirus (APRV) is currently the only member of this genus and was isolated from persistently infected Aedes pseudoscutellaris cells (AP61).
The morphology of purified APRV is very similar to that of the single-layered cypoviruses. Based on electron microscopy analysis, it was initially thought that APRV would represent a new cypovirus from mosquitoes. However, unlike the cypoviruses, APRV particles are not occluded in a crystalline matrix formed by a virus-encoded polyhedron protein, and full-length sequence analysis identifies only nine genome segments. The genetic distance between APRV and cypoviruses is consistent with their being members of different genera, similarto the relationship between oryzaviruses and fijiviruses (Table 1 Dinovernavirus).
Table 1 Dinovernavirus Amino acid identities between APRV and cypoviruses, fijiviruses and oryzaviruses
| APRV proteins | Lymantria dispar cypovirus 1 (Cypovirus) | rice ragged stunt virus (Oryzavirus) | Nilaparvata lugens reovirus (Fijivirus) | |||
|---|---|---|---|---|---|---|
| Protein (position) | % identity | Protein (position) | % identity | Protein (position) | % identity | |
| VP1 | P138 (768-1157) | 23 | P1 (963-1208) | 22 | ||
| VP2 | V2 (23-1206) | 26 | P4 (68-1030) | 23 | P165.9 (599–624) | [22] |
| VP3 | V1 (6-1302) | 21 | P3 (398-645) | 21 | P106.4 (339–698) | 21 |
| VP4 | NS1 (795-874) | 28 | P5 (727-789) | 31 | ||
| VP5 | V3 (38-1045) | 22 | P2 (589-1102) | 19 | ||
| VP6 | V4 (1-555) | 22 | NS7 (399-591) | 23 | P73.5 (399–591) | 23 |
| VP7 | ||||||
| VP8 | ||||||
| VP9 | NS35.2 (169–218) | 31 | ||||
Virion
Morphology
Purified APRV particles, isolated using an iodixanol gradient and analyzed by transmission electron microscopy, have the morphology typical of cores of turreted reoviruses (Figure 1 Dinovernavirus). In particular, the morphology of APRV appears similar to that observed for cypovirus particles, suggesting that APRV is also single-layered. The mean diameter of the particle is approximately 49–50 nm, with a central section that is 36–37 nm. Turrets were visible projecting from the particle surface. These also appear to have sections near the tip that can become detached (Figure 1 Dinovernavirus) in a manner similar to that observed previously with cypoviruses.
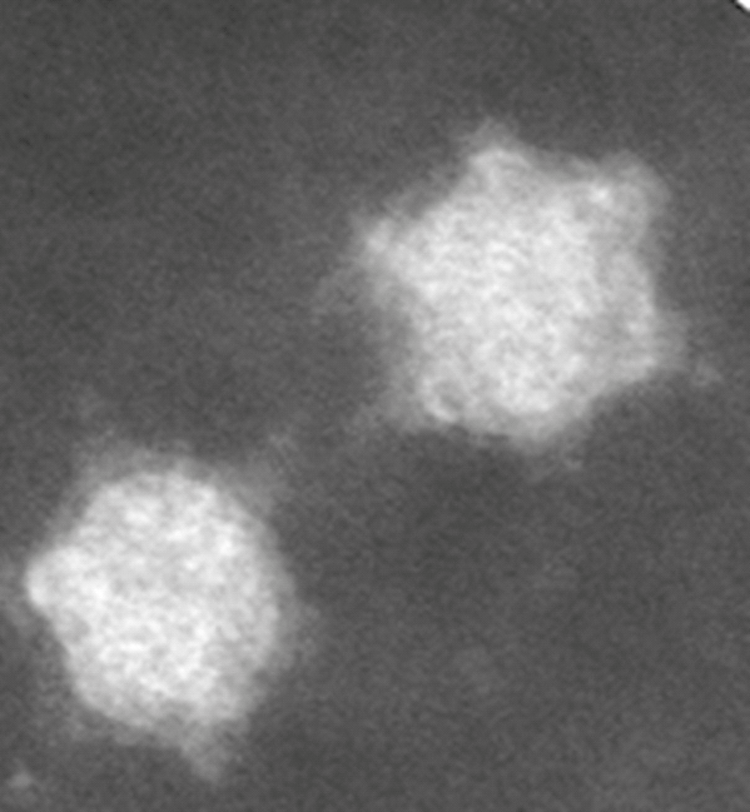 |
| Figure 1 Dinovernavirus Negative-contrast electron micrograph of particles of Aedes pseudoscutellaris reovirus purified using Iodixanol (Optiprep®) gradient. (Courtesy of H. Attoui). |
Physicochemical and physical properties
At 4 °C, APRV is stable for long periods, even as a cell culture lysate, so providing a convenient method of medium-term storage. Heating to 55 °C significantly reduces infectivity. The virion is stable upon treatment with freon, which can be used for purification of virions from cell lysate. Virion infectivity is not affected by treatment with 1% deoxycholate but is abolished by treatment with sodium dodecyl sulfate. Virions can be stored for long periods at −80 °C, and infectivity can be further protected by addition of 50% fetal calf serum. Infectivity is retained at pH values between 6 and 8. Between pH 4 and 5 or between pH 9 and 10, infectivity is reduced by a factor of 10. Virion morphology (observed by electron microscopy) is considerably distorted at pH values lower than 5 and virions are completely disrupted at pH values lower than 3.5.
Nucleic acid
The dinovernavirus genome consists of nine linear segments of dsRNA, which are numbered in order of reducing Mr, or increasing electrophoretic mobility during agarose gel electrophoresis (AGE). The genome comprises 23,355 bp, with segments ranging between 3,817 and 1,147 bp. Analysis of genomic RNAs by 1% AGE shows a 5-1-3 migration pattern (electropherotype) (Figure 2 Dinovernavirus).
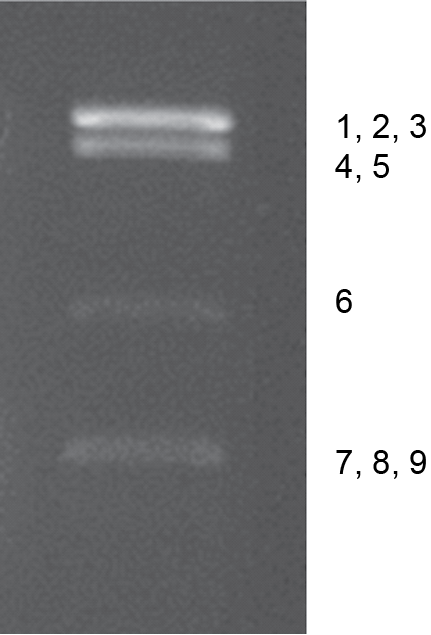 |
| Figure 2 Dinovernavirus Electrophoretic profile of genome of Aedes pseudoscutellaris reovirus (propagated in C6/36 cells) in 1% agarose gel. The segments comprising each band are indicated to the right. |
The positive-sense strands of all nine segments of the APRV genome have conserved sequences in the 5′- and 3′-non-coding regions (NCRs, 5′-AGUUA/UAAA/CA/C----------U/GUUnnnC/UnnA/UAGU-3′, where n = any nucleotide; Table 2 Dinovernavirus). Comparisons of these conserved termini with those of viruses in the genera Cypovirus, Oryzavirus and Fijivirus (Table 2 Dinovernavirus) shows that only the first three nucleotides in the 5′-NCR (AGU) are conserved between APRV, cypoviruses and Nilaparvata lugens reovirus (NLRV, species Fijivirus nilaparvatae). The 3′-termini of APRV differ from those of the cypoviruses, NLRV and rice ragged stunt virus (RRSV, species Oryzavirus oryzae).
In contrast to those of cypoviruses and fijiviruses, the first and last nucleotides of the APRV genome segments are complementary (A and U). The mean G+C content of the APRV genome is 34.4%, compared to 34.8% for NLRV, 44.7% for RRSV and 43% for cypoviruses.
Table 2 Dinovernavirus Conserved dinovernavirus terminal sequences (positive-sense strand) and comparison to those of cypoviruses, fijiviruses, oryzaviruses and idnoreoviruses
| Genus | Virus names | 5′-end | 3′-end |
|---|---|---|---|
| Dinovernavirus | Aedes pseudoscutellaris reovirus (APRV) | 5′-AGUUA/UAAA/CA/C | A/UAGU-3′ |
| Cypovirus | Lymantria dispar cypovirus 1(LdCPV1) | 5′-AGUAAA | GUUAGCC-3′ |
| Inachis io cypovirus 2 (IiCPV2) | 5′-AGUUU | GAGUUUGC-3′ | |
| Orgyia pseudotsugata cypovirus 5 (OpCPV5) | 5′-AGUUU/AU/A
| UUGC-3′ | |
| Antheraea mylitta cypovirus 4 (AmCPV4) | 5′-AAUCGACGACG | UAGAGC-3' | |
| Fijivirus | Nilaparvata lugens reovirus (NLRV) | 5′-AGU | GUUGUC-3′ |
| mal de Río Cuarto virus (MRCV) | 5′-AAGUUUUUU | GUC-3′ | |
| Fiji disease virus (FDV) | 5′-AAGUUUUUU | GUC-3′ | |
| rice black streaked dwarf virus (RBSDV) | 5′-AAGUUUUUU | GUC-3′ | |
| Oryzavirus | rice ragged stunt virus (RRSV) | 5′-GAUAAA | GUGC-3′ |
| Idnoreovirus | Operophtera brumata idnoreovirus (ObIRV) | 5′-AAA/CA/UAA | AGGUU-3′ |
Proteins
See Table 3.Dinovernavirus.
Genome organization and replication
APRV proteins were inferred from the ORFs in the dsRNA segments. Each segment encodes a single protein (Table 3 Dinovernavirus).
Table 3 Dinovernavirus Genome segments and protein products of Aedes pseudoscutellaris reovirus
| Genome segment | bp | Protein nomenclature | Protein mass (kDa) | Structure/putative function |
|---|---|---|---|---|
| Seg1 | 3817 | VP1 | 134 | |
| Seg2 | 3752 | VP2 | 143 | RNA-directed RNA polymerase (RdRP) |
| Seg3 | 3732 | VP3 | 136 | |
| Seg4 | 3375 | VP4 | 116 | |
| Seg5 | 3227 | VP5 | 121 | Possible guanylyltransferase |
| Seg6 | 1775 | VP6 | 62 | |
| Seg7 | 1171 | VP7 | 39.4 | |
| Seg8 | 1151 | VP8 | 39.8 | |
| Seg9 | 1147 | VP9 | 32 |
APRV persistently infects the Aedes pseudoscutellaris AP61 cell line, with an estimated 6–8 particles per cell. Treatment of cells with 2-aminopurine showed a 10-fold increase in the number of viral particles, as shown by qPCR. The virus replicates to high titres in C6/36 Aedes albopictus cells with over 40% of progeny liberated into the culture medium.
APRV does not replicate in a range of mammalian cells including L929 (mouse fibroblast), BHK-21 (hamster kidney), Vero (monkey kidney), BGM (monkey kidney), Hep-2 (human cervix cancer) and MRC-5 (human lung fibroblasts). RNA extracts of blood from inoculated Balb/c mice remained negative for virus RNA by RT-PCR from 0 to 12 days post-injection.
Biology
APRV was isolated from persistently infected Aedes pseudoscutellaris AP61 cells. AP61 cells collected from different sources contain the same APRV strain. Virus-like particles were also identified in the original AP61 line by electron microscopy. Care should be taken when using AP61 cells to propagate any other mosquito-borne arboviruses, particularly dsRNA viruses such as orbiviruses and seadornaviruses, as these readily become contaminated with APRV. Upon infection with other dsRNA viruses, the dormant or persistent state of APRV is activated to a productive infection, releasing APRV into the cell culture supernatant, along with other mosquito-borne dsRNA viruses used to infect the cells.

