Family: Spinareoviridae
Genus: Fijivirus
Distinguishing features
Fijivirus particles have a double-layered, icosahedral structure, with a spherical rather than angular appearance and short surface spikes (A spikes) on each of the 12 vertices of the icosahedron. The outer shell is fragile and easily breaks down, leaving the inner shell bearing 12 B spikes. There are 10 genome segments. The viruses replicate in delphacid planthoppers. Nilaparvata lugens reovirus (NLRV) has the above properties but replicates only in insects, whereas other fijiviruses can also replicate in phloem cells of susceptible plants of the families Gramineae (in which they induce small tumors or enations), Cyperaceae or Liliaceae (Li et al., 2012). Characteristic of the genus is the low G+C content of the genomic RNAs, mostly around 34–36%.
Virion
Morphology
Virions are double-layered, spherical particles, 65–70 nm in diameter with A spikes (about 11 nm in length and breadth) at the 12 vertices on the icosahedra (Figure 1 Fijivirus, left). Unless pre-fixed, virions readily break down in vitro to give cores, about 55 nm in diameter, with 12 B spikes, about 8 nm long and 12 nm in diameter (Figure 1 Fijivirus, right). Some treatments (shaking with butan-1-ol or incubation with 1.9 M MgCl2) produce smooth subcores (Figure 1.Fijivirus, center).
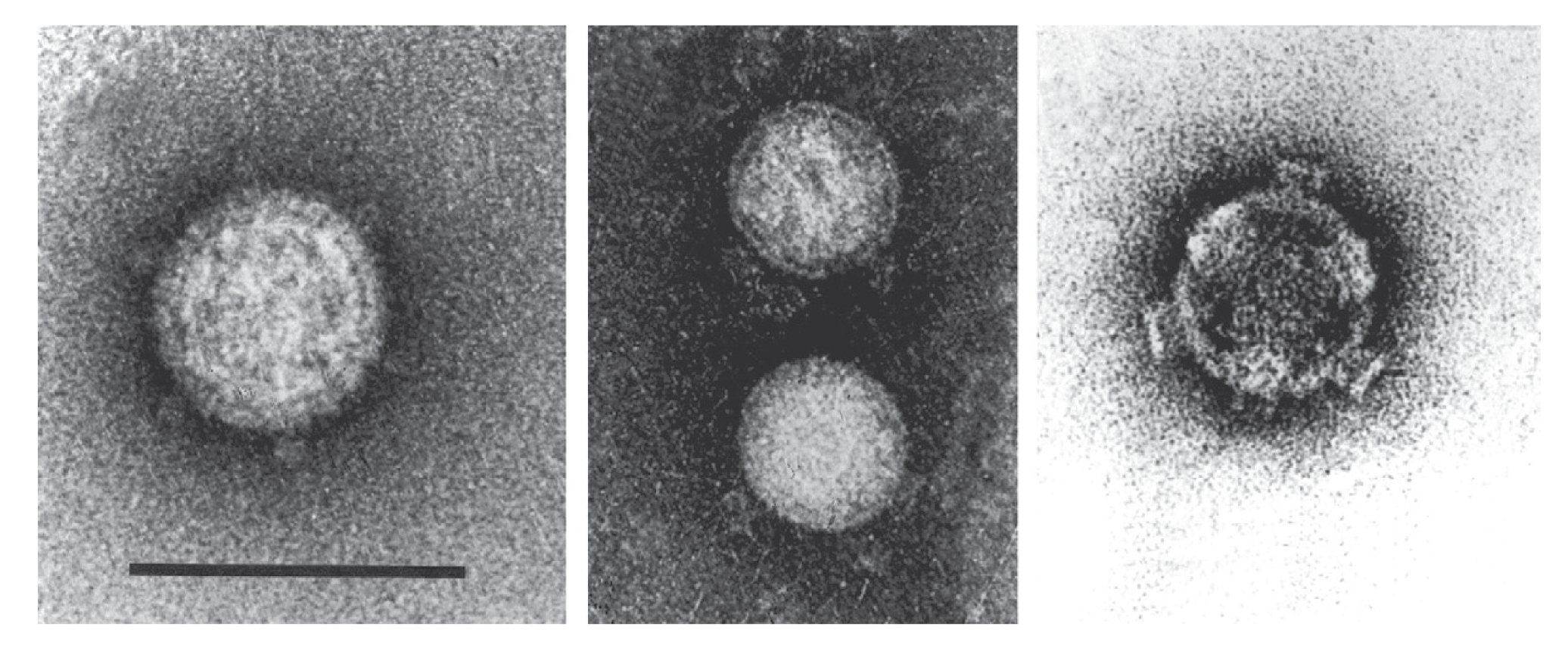 |
| Figure 1 Fijivirus (Left) Negative-contrast electron micrograph of maize rough dwarf virus virions stained with uranyl acetate showing “A” spikes; (center) smooth subcores derived on staining virion with neutral phosphotungstate; (right) “B” spikes on virus-derived cores stained with uranyl acetate (courtesy of R. G. Milne). The bar represents 100 nm. |
Nucleic acid
Fijiviruses have 10 dsRNA segments that are numbered in order of increasing electrophoretic mobility during polyacrylamide gel electrophoresis. Some segments do not migrate in order of their Mr and may migrate in a reverse order during (1%) agarose gel electrophoresis (AGE). Examples include segments 2 and 3 of NLRV and mal de Río Cuarto virus (MRCV) and segments 8 and 9 of oat sterile dwarf virus (OSDV). The conserved terminal sequences are shown in Table 1 Fijivirus. Within the genus, only the 3′-terminal sequence …GUC-3′ is conserved. Adjacent to the conserved terminal oligonucleotide sequences, each genome segment possesses inverted repeats of several base pairs, similar to those in phytoreovirus and oryzavirus RNAs, although the sequences involved differ in these other genera. Characteristic of the genus is the low G+C content of the genomic RNAs, mostly around 34–36%. The lengths and groupings of the 10 dsRNA species are characteristic and distinctive for the five groups of fijiviruses that are recognized (see Biology).
Table 1 Fijivirus Conserved terminal sequences (positive-sense strand) of viruses of the genus Fijivirus.
| Virus species | Virus name | 5′-end | 3′-end |
|---|---|---|---|
| Fijivirus fijiense | Fiji disease virus (FDV) | 5′-AAGUUUUU | CAGC(A/U)(A/G)(A/G)(C/U)GUC-3′ |
| Fijivirus zeae | maize rough dwarf virus (MRDV) | 5′-AAGUUUUUU | CAGCU(A/G)N(C/U)GUC-3´ |
| Fijivirus cuartoense | mal de Río Cuarto virus (MRCV) | 5′-AAGUUUUU | CAGCU(A/G)(A/U)(C/U)GUC-3′ |
| Fijivirus nilaparvatae | Nilaparvata lugens reovirus (NLRV) | 5′-AGU | GUUGUC-3′ |
| Fijivirus alporyzae | rice black streaked dwarf virus (RBSDV) | 5′-AAGUUUUU | CAGCUNN(C/U)GUC-3′ |
| Fijivirus boryzae | southern rice black-streaked dwarf virus (SRBSDV) | 5′-AAGUUUUU | CAGCUGAUGUC-3′ |
| Fijivirus avenae | oat sterile dwarf virus (OSDV) | 5′-AACGAAAAAAA | UUUUUUUUAGUC-3′ |
Proteins
Six polypeptides, numbered respectively I to VI (139, 126, 123, 111, 97 and 64 kDa), can be detected by SDS PAGE of purified maize rough dwarf virus (MRDV) virions. The B-spiked cores contain peptides I, II and III, while the smooth core contains peptides I and II. The B spikes should therefore be composed of peptide III. Peptides IV–VI form the outer capsid.
Three major proteins (130, 120 and 56 kDa) and three minor ones (148, 65 and 51 kDa) can be detected by SDS PAGE of purified virions of rice black streaked dwarf virus (RBSDV). The 120 kDa protein is the B spike protein. Smooth subcore particles consist of 148, 130 and 65 kDa proteins. The 56 kDa protein is the major component of the outer capsid shell and the 51 kDa protein is a partial degradation form. In NLRV virions, three major proteins (140, 135 and 65 kDa), three intermediate (160, 110 and 75 kDa), and one minor protein (120 kDa) can be resolved. The 135 kDa protein is the B spike. The 65 kDa protein is the major component of the outer capsid shell and the 140 kDa protein is the major core protein. In addition to the above structural proteins, there is an A spike but its protein has not yet been identified (Isogai et al., 1998).
RBSDV encodes at least six putative structural proteins (P1, P2, P3, P4, P8, and P10) and six putative non-structural proteins (P5, P6, P7-1, P7-2, P9-1, and P9-2). Among the structural proteins, P1 contains the characteristic sequence motifs of an RNA-directed RNA polymerase (RdRP); P2 and P4 are the major core and spike protein, respectively; P3 is a putative capping enzyme; and P8 and P10 are a core and major outer capsid proteins, respectively.
Crystallographic analysis of RBSDV P9-1 reveals that this protein multimerizes giving rise to dimers, tetramers and cylindrical octamers with internal pores (Akita et al., 2012). Interestingly, P9-1 preferentially binds to ssRNA in its octamer form and the electropositive amino acids among residues 25–44 that are important for RNA binding map to the octamer central internal pore (Wu et al., 2013).
Genome organization and replication
Genome organizations and coding assignments of fijiviruses are summarized in Table 2 Fijivirus. Most of the genome segments are monogenic. Some segments possess two ORFs but expression of the second ORF has not been demonstrated in vivo in insect or plant cells. For viruses other than NLRV, replication occurs in the cytoplasm of phloem-related cells in association with viroplasm. NLRV does not have a counterpart to the ORF2 present in Fiji disease virus (FDV) Seg7 (and the corresponding segments of other plant-infecting fijiviruses), and this may reflect its inability to replicate in plant hosts.
During infection of most (possibly all) fijiviruses, virus-containing tubules about 90 nm in diameter accumulate in the cytoplasm. Sometimes these are incompletely closed and form scrolls. Such virus-associated tubules or scrolls are composed of non-structural proteins and facilitate viral spread in insect vectors. Southern rice black-streaked dwarf virus (SRBSDV) uses the tubules composed by non-structural prtotein P7-1 as a vehicle for rapid spread of virions through basal lamina from midgut epithelium toward visceral muscle tissues of its vector, the white-backed planthopper (Sogatella furcifera (Horváth, 1899)) (Jia et al., 2014). Replication and assembly of progeny virions of fijiviruses are thought to occur within the viroplasms composed of granular and filamentous regions during viral infection of the plant host or insect vector. The viroplasm induced by SRBSDV infection consists of a granular region, where viral RNAs and non-structural proteins P6 and P9-1 have accumulated, and a filamentous region, containing viral RNAs, progeny cores, viral particles, and non-structural proteins P5 and P6. Non-structural proteins P5, P6, and P9-1 are collectively required for the genesis and maturation of the filamentous and granular viroplasm matrix induced by SRBSDV infection (Mao et al., 2013).
Table 2 Fijivirus Genome segments and protein products of rice black streaked dwarf virus (Hubei isolate) isolated from maize (Wang et al., 2003).
| Genome segment | bp | Protein Mr predicted (kDa) | Location (function)* | Homologous segment in other members of the genus | |||||
| MRCV | MRDV | OSDV | SRBSDV | FDV | NLRV | ||||
| Seg1 | 4501 | 168.5 | RdRP | 1 | 1 | na | 1 | 1 | 1 |
| Seg2 | 3813 | 141.3 | Major core protein | 3 | 2 | na | 2 | 3 | 3 |
| Seg3 | 3572 | 131.8 | Possible guanylyltransferase | 4 | 3 | na | 4 | 4 | 4 |
| Seg4 | 3617 | 135.2 | Outer shell, possible B spike | 2 | 4 | na | 3 | 3 | 2 |
| Seg5 | 3164 | 106.9 | Non-structural protein, filamentous viroplasm matrix protein | 5 | 5 | na | 5 | 5 | 5 |
| Seg6 | 2645 | 89.6 | Non-structural protein, viroplasm matrix protein, RNA-silencing suppressor | 6 | 6 | na | 6 | 6 | 6 |
| Seg7 | 2193 | 40.9 | Non-structural protein, tubular protein | 7 | 7 | 7 | 7 | 7 | 10** |
| 36.7 | Unknown | ||||||||
| Seg8 | 1936 | 68 | Core protein, possible NTP-binding | 8 | 8 | 9 | 8 | 8 | 7 |
| Seg9 | 1900 | 39.9 | Non-structural protein, granular viroplasm matrix protein | 9 | 9 | 10 | 9 | 9 | 9 |
| 24.2 | Non-structural protein | ||||||||
| Seg10 | 1801 | 63.0 | Major outer capsid protein | 10 | 10 | 8 | 10 | 10 | 8 |
* The probable function of some of the proteins has been deduced from the equivalent genome segment of other virus species.
** Genome Seg10 of NLRV does not contain a second ORF.
Biology
All the plant-infecting fijiviruses induce hypertrophy of the phloem (both expansion and multiplication of cells), leading to vein swellings and sometimes galls (enations or tumors) derived from phloem cells, especially on the backs of leaves. MRDV in maize induces longitudinal splitting of the roots. Other effects include the suppression of flowering, plant stunting, increased production of side shoots, and induction of a dark green coloring. In insect hosts, no particular tissue tropism or severe disease is recognized. Viruses are transmitted propagatively by delphacid planthoppers (Hemiptera: Delphacidae, e.g., Perkinsiella, Laodelphax, Toya, Sogatella, Javesella, Ribautodelphax, Dicranotropis, Delphacodes, Sogatella, and Unkanodes). Following virus acquisition from infected plants, the latent period is about two weeks, and leads to a lifelong capacity for virus transmission to plants. No transovarial or seed transmission of virus has been identified. Mechanical transmission from plant to plant can be demonstrated only with difficulty. Virus is spread by offsets in vegetatively propagated crops (e.g., pangola grass and sugarcane). Viruses can overwinter in diapausing planthoppers, in certain weed species and in autumn-sown cereals.
Generally, fijiviruses are widespread in nature, although they are apparently absent from North America and have not been reported from Africa or confirmed from India. Fiji disease virus (FDV) has been reported from Australia and the Pacific islands. RBSDV occurs in Japan, Korea and China. SRBSDV occurs in China, Vietnam, and Japan (Zhou et al., 2013). Pangola stunt virus (PaSV) occurs in northern countries of South America, Oceania, Taiwan and northern Australia, and OSDV occurs in northern Europe. Garlic dwarf virus (GDV) has been found only in southern France. MRDV is found in areas bordering the northern and eastern Mediterranean. MRCV occurs in Argentina. NLRV was found in Nilaparvata lugens (Stål, 1854) planthoppers which can be found in south-east Asia. Experimentally, NLRV also infects Laodelphax striatellus (Fallén, 1826) grasshoppers. There is no evidence that NLRV can multiply in rice plants, a natural host of N. lugens planthoppers, but the virus is transmitted from grasshopper to grasshopper through contaminated rice plants and moves through the phloem or xylem of rice plants once injected by the viruliferous hoppers.
Antigenicity
Some proteins of the viruses in group 2 (MRCV, MRDV, Pangola stunt virus (PaSV), RBSDV and SRBSDV) are distantly related but homologous. Proteins from viruses of other species in the genus are serologically unrelated.
Species demarcation criteria
Of the nine fijivirus species, members of five (collectively referred to as group 2) are relatively closely related to one another (Figure 2 Fijivirus). Further information about these viruses may eventually necessitate a revision of their species status. In particular, MRDV and RBSDV may be considered sufficiently closely related to constitute a single species.
 |
| Figure 2 Fijivirus Phylogenetic relationships of fijiviruses major outer capsid proteins. Amino acid sequences were aligned using MUSCLE (Edgar 2004) and the tree constructed in MEGA7 (Kumar et al., 2016) using the neighbour-joining method and the p-distance algorithm with pairwise deletion. Bootstrapping values (1000 replicates) above 70 are shown. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
The conserved terminal sequences of genome segments do not differ greatly between fijiviruses (Table 1 Fijivirus). In addition to the other general criteria used throughout the family, members of a species in the genus Fijivirus may be identified by:
- Sequence analysis: members of different species usually have <40% amino acid identity in comparisons of proteins corresponding to those encoded by RBSDV segments 7, 8, 9 and 10. In comparisons among the genome segments coding for the major capsid protein, viruses from different groups have <55% nucleotide identity (but identities are much higher within the group 2, vide supra).
- Sequences of the 5´ and 3´ terminal ends.
- Serological cross-reactions: viruses in different groups do not cross-react; those in group 2 do so to a limited extent that is dependent on the proteins being compared.
- The identity or family of the plant host species (if any) together with the insect vector and its host.Historically, viruses assigned to the genus Fijivirus were divided into five groups based on the host and phylogeny. Group one solely contains Fiji disease virus (FDV); group 2 contains four closely related members: maize rough dwarf virus (MRDV), mal de Río Cuarto virus (MRCV), rice black streaked dwarf virus (RBSDV) and southern rice black-streaked dwarf virus (SRBSDV); groups 3, 4 and 5 only contain one member each, oat sterile dwarf virus (OSDV), garlic dwarf virus (GDV) and NLRV respectively. Members of groups 1, 2 and 3 replicate in plants of the family Gramineae. Members of groups 2 also replicate in plants of the family Cyperaceae. GDV replicates in plants of the family Liliaceae. NLRV does not replicate in plants. NLRV replicates in the planthopper Nilaparvata lugens, but does not replicate in plants.

