Family: Sedoreoviridae
Genus: Seadornavirus
Distinguishing features
The seadornavirus genome consists of 12 segments of dsRNA. During replication, virions are found in the cell cytoplasm. Viruses are transmitted to vertebrate hosts by mosquito vectors.
Virion
Morphology
Particles are non-enveloped with a diameter ranging from 60−70 nm having two concentric capsid shells with a core that is about 40–50 nm in diameter. Electron microscopic studies, using negative-staining have shown that particles have a well-defined surface capsomeric structure and icosahedral symmetry (Figure 1.Seadornavirus).
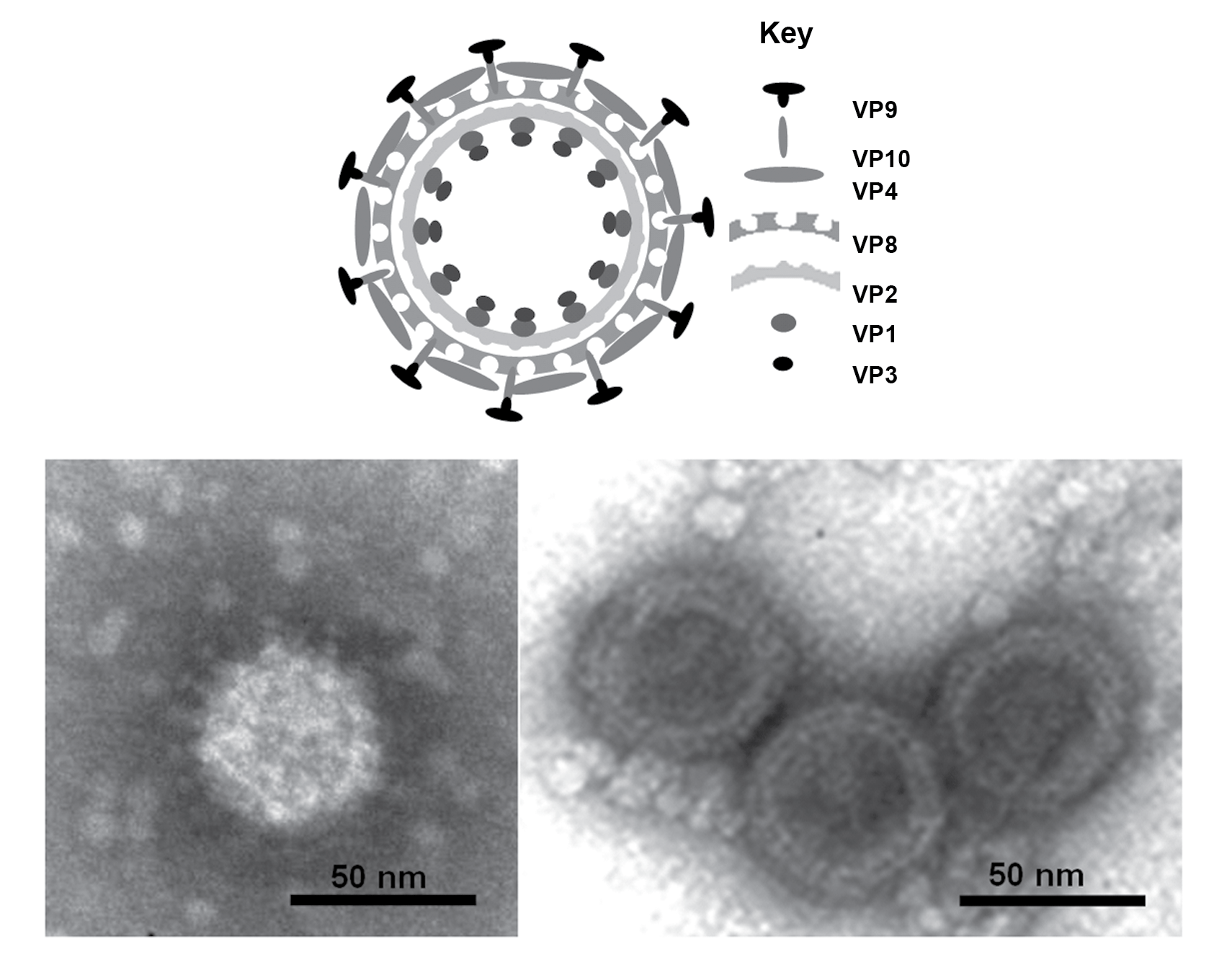 |
| Figure 1. Seadornavirus (Top) Diagram of Banna virus virion structure, constructed using data from biochemical analyses, electron microscopy and X-ray crystallography. (Bottom) Negative-contrast electron micrograph of virions: (left hand side) full particles showing multiple protein spikes; (right hand side) double layered cores. (Courtesy of H. Attoui). |
Physicochemical and physical properties
The buoyant density of virions in CsCl is 1.36 g cm−3. Virions are stable around neutral pH but lose infectivity at pH 3.0. At 4 °C, virions are stable for long periods, even non-purified in cell culture lysate, which provides a convenient method for medium-term storage. Heating to 55 °C considerably decreases viral infectivity. Seadornaviruses are stable upon treatment with freon, which could be used for purification of viral particles from cell lysate. Viral infectivity is abolished by treatment with sodium dodecyl sulfate. Virions can be stored for long periods at −80 °C, and infectivity is further protected by addition of 50% fetal calf serum.
Nucleic acid
The genome consists of 12 dsRNA segments that are numbered in order of reducing Mr, or increasing electrophoretic mobility during agarose gel electrophoresis. The genome comprises approximately 21,000 bp, with segment ranging from 3,747 to 862 bp. The RNA genome segments of Banna virus (BAV), Kadipiro virus (KDV) and Liao ning virus (LNV) migrate as groups of 6-6, 6-5-1 and 6-2-3-1, respectively during 1% agarose gel electrophoresis (AGE) (Figure 2.Seadornavirus). These patterns are thought to be characteristic for each virus species.
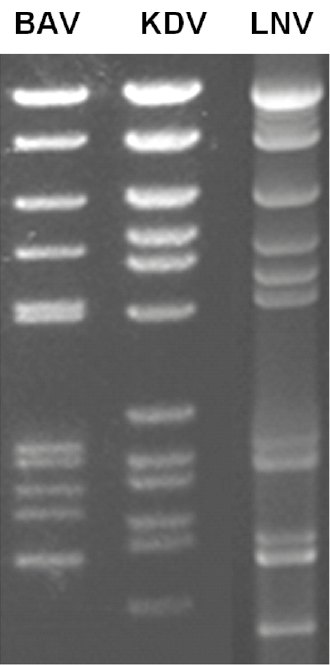 |
| Figure 2. Seadornavirus Migration patterns (electropherotypes) of genome segments from isolates of Banna virus (BAV), Kadipiro virus (KDV) and Liáo níng virus (LNV) in 1% agarose gel. Electropherotypes are characteristic of each virus species. |
Full-genome sequence analysis of Banna virus isolates has identified two genotypes based on Seg7 and Seg9 sequences that correlate with virus serotype. Genotype A includes isolates BAV-Ch (China) and BAV-In6423 (Indonesia), whereas genotype B includes BAV-In6969 (Indonesia) and BAV-In7043 (Indonesia). The proteins translated from Seg7 and Seg9 have 72% and 54% amino acid identity between genotypes A and B respectively, whereas all of the other segments appear to be more conserved, having 83% to 98% identity (Attoui et al., 2005). Two genotypes of LNV have also been identified that correlate with virus serotype. Amino acid identity between these two genotypes in the cell attachment outer capsid protein VP10 is 81%.
Much lower levels of amino acid sequence identity are detected between homologous proteins of members of different species, being 24–42% between BAV and KDV, 18–41% between BAV and LNV, and 21–42% between LNV and KDV. In each case the highest levels of amino acid identity were detected in the viral RNA-directed RNA polymerase (RdRP, VP1). Phylogenetic relationships for the RdRP of BAV, KDV and LNV are shown in Figure 3.Seadornavirus.
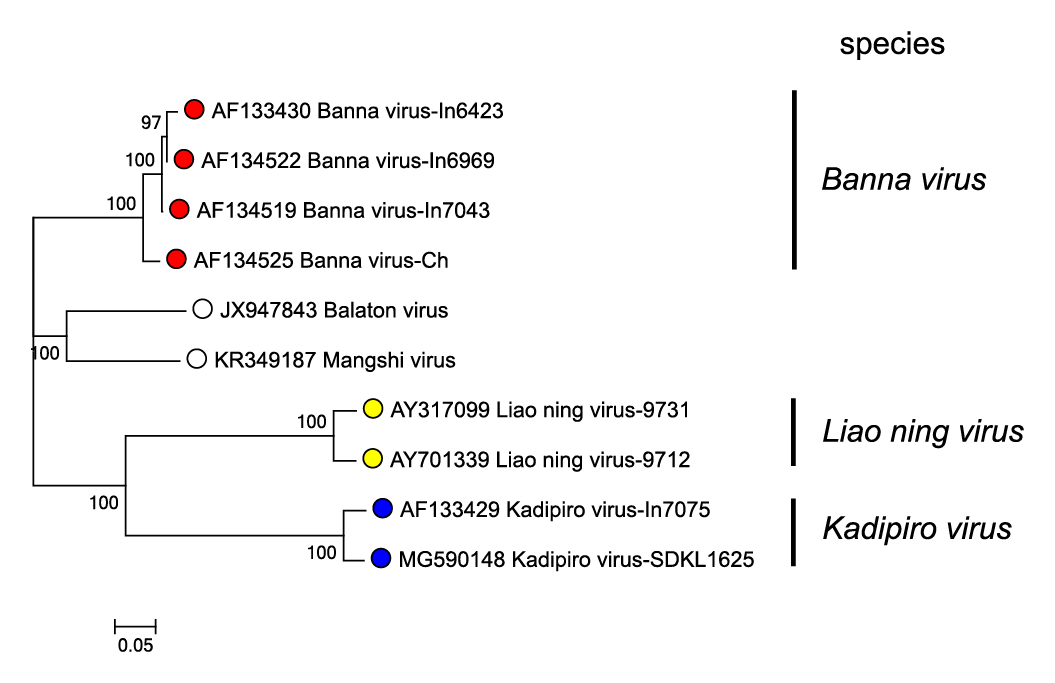 |
| Figure 3.Seadornavirus Phylogenetic relationships between RdRP (VP1) amino acid sequences of members of different seadornaviruses species. Sequences were aligned using MUSCLE (Edgar 2004) and the tree constructed in MEGA7 (Kumar et al., 2016) using the neighbour-joining method and the p-distance algorithm with pairwise deletion. Bootstrapping values (1000 replicates) above 70 % are shown. The unclassified viruses Balaton virus and Mangshi virus are likely to represent novel species. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
The genome segments of BAV and KDV have a G+C content that varies between 37% and 39%.The 5′-NTRs are 17 to 111 nucleotides long while the 3′-NTRs are 76 to 281 nucleotides long. The NTRs of seaodornaviruses contain conserved base pairs at both termini (positive-sense strand 5′-GU…GAC-3′) (Table 1.Seadornavirus). Fully functional and infectious virions have been recovered by the introduction of all twelve mRNAs into BSR cells.
Table 1.Seadornavirus Conserved seadornavirus terminal sequences (positive-sense strand)
|
Virus species |
Virus name |
5′-end |
3′-end |
|
Banna virus |
Banna virus (BAV) |
5′-GUAUA/UA/UAAA/UA/UU |
A/GCC/UGAC-3′ |
|
Kadipiro virus |
Kadipiro virus (KDV) |
5′-GUAGAAA/UA/UA/UU |
AA/CC/UGAC-3′ |
|
Liao ning virus |
Liao ning virus (LNV) |
5′-GUUAUA/UA/UA/U |
A/CU/CCGAC-3′ |
Proteins
Native proteins of BAV particles have 138been characterized by mass spectrometry analysis and by radiolabeling of infected cells. Their putative functions are indicated in Table 2.Seadornavirus.
Table 2.Seadornavirus Genome segments and protein products of Banna virus (BAV-In6423)
|
Genome segment |
bp |
Protein [copy number per particle] |
Protein mass (kDa)* |
Structure/function |
|
Seg1 |
3747 |
VP1(Pol) [24] |
137 |
RdRP (transcription complex) |
|
Seg2 |
3048 |
VP2(T2) [120] |
108 |
Inner layer of core, T2 protein |
|
Seg3 |
2400 |
VP3(Cap) [12] |
82 |
Capping enzyme (transcription complexe) |
|
Seg4 |
2038 |
VP4 [333] |
64 |
Outer coat protein |
|
Seg5 |
1716 |
VP5-NS [0] |
56 |
Non-structural |
|
Seg6 |
1671 |
VP6-NS [0] |
48 |
Non-structural |
|
Seg7 |
1136 |
VP7-NS [0] |
35 |
Protein kinase |
|
Seg8 |
1119 |
VP8 [780] |
32 |
T13, outer layer of core |
|
Seg9 |
1101 |
VP9 [310] |
31 |
Outer coat cell attachment protein |
|
Seg10 |
977 |
VP10 [310] |
29 |
Stalk base for VP10 |
|
Seg11 |
867 |
VP11-NS [0] |
21 |
Non-structural |
|
Seg12 |
862 |
VP12-NS [0] |
24 |
Non-structural dsRNA-binding |
* Calculated from nucleotide sequences
Purified BAV-Ch virions contain seven structural proteins, each of which co-migrates with one of the radio-labeled proteins from infected cells (VP1, VP2, VP3, VP4, VP8, VP9 and VP10). Only five of these proteins are also detected in cores, indicating that the outer coat (like those of the non-turreted orbiviruses and rotaviruses) is composed of two proteins (VP4 and VP9). Analyses of BAV-Ch structural protein sequences by mass spectrometry confirms the identity of the core (VP1, VP2, VP3, VP8, VP10) and outer capsid components (made of VP9 and VP4), demonstrating that VP5, VP6, VP7, VP11 and VP12 are non-structural proteins.
SDS-PAGE analysis of purified [35S] methionine labeled BAV-Ch particles shows that VP2 and VP8 are the two most abundant proteins of the BAV core. The lower relative abundance and higher molecular weight of VP2 identifies it as the subcore-shell T2 protein (equivalent to VP3 of bluetongue virus (BTV) and VP2 of rotavirus A). In contrast, VP8 is smaller and more abundant, identifying it as the core-surface T13 protein. VP8 and VP2 have a molar ratio of 6.5 in purified BAV-Ch particles, identical to the ratio of 6.5previously detected between the subcore and core-surface proteins of both BTV and rotaviruses. On this basis, the numbers of the VP8 and VP2 molecules in the BAV core are assumed to be 780 and 120 per particle, respectively, allowing the average copy number of the other protein components of virions or cores to be calculated: 24 copies for VP1(Pol), 12 copies for VP3(Cap), 333 copies for VP4, and 310 copies for both VP9 and VP10.
The structure of the BAV outer-capsid protein VP9 determined by X-ray crystallography at 2.6 Å resolution, reveals a trimeric molecule, held together by an N-terminal helical bundle, reminiscent of coiled-coil structures found in fusion-active proteins such as the gp41 ofhuman immunodeficiency virus (Retroviridae) The major domain of VP9 contains stacked β sheets with marked structural similarities to the rotavirus A receptor binding protein VP8. Anti-VP9 antibodies neutralize viral infectivity, and, remarkably, pretreatment of cells with trimeric VP9 increases viral infectivity, indicating that VP9 is involved in virus attachment to cell surface and subsequent internalization. Sequence similarities between BAV VP10 and the VP5 portion of rotavirus VP4, suggest that the receptor binding and internalization apparatus, which is a single gene product activated by rotavirus proteoloysis, is the product of two separate genome segments in BAV.
Genome organization and replication
The 12 genome segments of seadornaviruses are all monogenic (Table 2.Seadornavirus). Seadornavirus isolates replicate in mosquito cell lines, and considerable amounts of virus (over 40% of progeny) are liberated into the culture medium prior to cell death and gross CPE, which usually occurs by 40 hours post infection with BAV, and 72 hours post infection with KDV. BAV also replicates in BSR cells (a clone of BHK-21). LNV is the only seadornavirus known to replicate in a wide variety of mammalian cells, including primary cell cultures.
Host–cell protein synthesis shut-off starts at 2 hours after BAV infection of Aedes albopictus mosquito C6/36 cells, and the shut-off is complete by 6 hours post infection. [35S]-methionine added to C6/36 cell cultures at 6 hours post infection was incorporated almost exclusively into 12 protein bands (resolved by SDS PAGE), which are thought to represent the different viral proteins (one protein per genome segment). Most of these proteins have apparent molecular masses that agree with the theoretical masses predicted by sequence analysis of the viral genome. The only exception is VP7, which migrates more slowly than expected.
Large electron-dense structures occur in the cytoplasm of BAV-Ch infected cells, which correspond to the viral inclusion bodies (VIB) thought to be the main site of replication and particle assembly of other reoviruses. Particles (ca. 50 nm in diameter) with a smooth surface are detected mainly at the periphery of the VIB, although some particles occur within the VIB matrix. Virion are also detected within large vacuoles dispersed throughout the infected cell cytoplasm. These vacuoles contain multiple double-layered vesicles, lined with viral particles (ca. 50 nm in diameter) at their inner surface and it is possible that this reflects some involvement of cellular membrane structures or organelles in virus morphogenesis, transport or replication (as previously reported for rotaviruses). Virion entry into cells by endocytosis is suggested by the detection of virions in pits at the cell surface. Virions are also observed near the cell membrane, appearing to bud from the cell surface.
Biology
Seadornaviruses have been isolated from humans and mosquitoe vectors. The mosquitos that have been implicated include those of the species Culex vishnui Baisas, 1938, C. fuscocephalus Theobald, 1907, Anopeles vagus Dönitz, 1902, Anopheles aconitus Dönitz, 1902, Anopheles subpictus (Grassi, 1899) and Aedes dorsalis (Meigen, 1830). Experimentally, seadornaviruses have been found to replicate in adult mice, being detected in infected mouse blood 3 days post infection. LNV kills adult Balb/c mice causing a hemorrhagic syndrome. BAV was first isolated from the serum and cerebrospinal fluids of human patients showing neurological manifestations. The pathology provoked by BAV is only poorly described. KDV has only been isolated from mosquitoes.
BAV and KDV occur in tropical and subtropical regions, where other mosquito-borne viral disease especially Japanese encephalitis and dengue have been reported as endemic. Despite the isolation of BAV from infected human patients, no surveys have been reported concerning the detection and prevalence of antibodies to these viruses in human sera. LNV has been isolated from mosquitoes.
Antigenicity
BAV from China, Vietnam and Indonesia, KDV from China and Indonesia and LNV from China are classified as members of three different species and show little cross-reaction in neutralization tests.
Species demarcation criteria
In addition to the other general criteria used throughout the family, members of a species in the genus Seadornavirus may be identified by:
- Relatively efficient cross-hybridization of conserved genome segments under high stringency conditions (>74% identity).
- Serological comparisons by neutralization assays. There is no cross-neutralisation or cross-reactivity between members of the species Banna virus and Kadipiro virus, but lack of cross-neutralisation can also be observed between different genotypes of virus within the species Banna virus.
- Sequence analysis: In the conserved Seg12, viruses within the same species will normally have >89% nucleotide identity. The RdRPs (the most conserved protein, encoded by Seg1) of isolates of different species have <50% amino acid identity (Figure 3.Seadornavirus).
The sequence relationships between different members of the genus are illustrated in the RdRP based phylogenetic tree for Banna virus (BAV), Kadipiro virus (KDV) and Liao ning virus (LNV) (Figure 3.Seadornavirus). Within a single species, amino acid identity is >94%. Between viruses of different species, aa identity ranges from 40 to 43%.
Related, unclassified viruses
|
Virus name |
Accession |
Abbreviation |
|
Balatone virus |
Seg1: JX947843; Seg3: JX947844; |
BALV |
|
Mángshì virus |
Seg1: KR349187; Seg2: KR349188; |
MANGV |
Virus names and virus abbreviations are not official ICTV designations.
Balatone virus was isolated in Hungary while Mángshì virus was isolated in China.

