Family: Sedoreoviridae
Genus: Rotavirus
Distinguishing features
Rotaviruses infect only vertebrates and are transmitted by a fecal–oral route. When viewed by negative-contrast electron microscopy (Figure 1.Rotavirus), virions have a wheel-like appearance from which the genus derives its name (Latin rota, “wheel”). The triple-layered capsid encloses a genome of 11 linear dsRNA segments and is formed in a unique morphogenic pathway, which involves acquisition of a transient lipid envelope during budding of immature particles into the endoplasmic reticulum (ER).
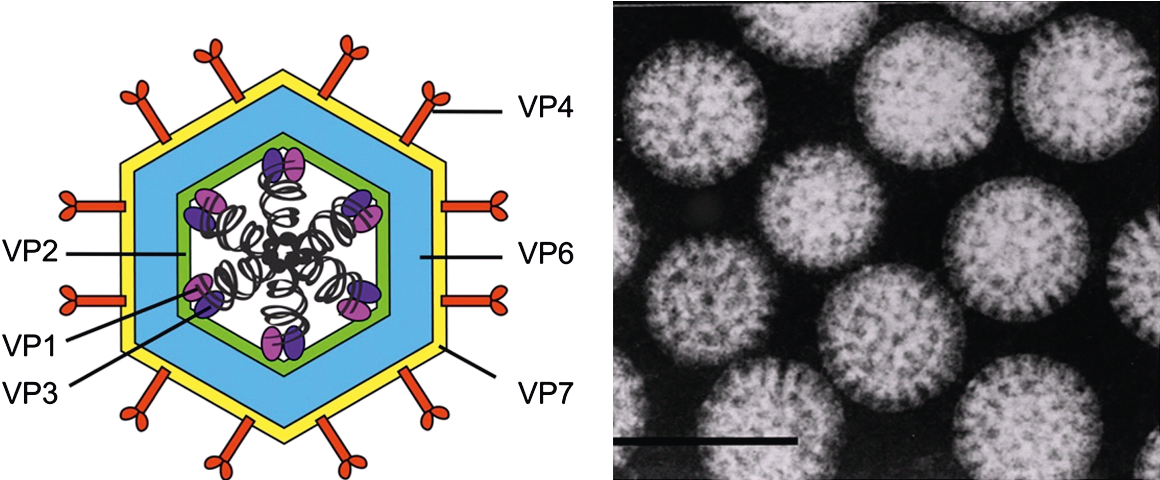 |
| Figure 1. Rotavirus Rotavirion morphology. (Left panel) Cartoon representation of a rotavirus triple-layered particle. Black corkscrews represent segments of genomic dsRNA and proteins are labelled. The precise locations of VP1 and VP3 are not known. (Right panel) Electron micrograph of rotavirus particles viewed by negative-staining. Bar represents 100 nm. (Provided by B. V. V. Prasad.) |
Virion
Morphology
The data for simian rotavirus A, strain SA11 (RVA-SA11) represent a paradigm for other viruses within the genus. The mature infectious virion has an overall diameter of about 100 nm, is made up of three concentric protein layers and lacks a lipid-containing envelope (Figure 1.Rotavirus) (Flewett et al., 1974). The detailed topology of these layers and their protein components has been revealed using cryoEM (Jayaram et al., 2004), followed by image processing of viral and subviral particles, as well as of virus-like particles formed using recombinant baculoviruses that express specific rotavirus structural proteins (Figure 2.Rotavirus). The innermost layer of the virion, composed of VP2, is about 3.5 nm thick. This layer is comparable to the internal capsid layer of members of other genera within the order Reovirales (e.g., the VP3 (T2) layer of the orbiviruses). The VP2 layer (T=1; 60 asymmetric dimers of VP2) surrounds the genomic dsRNAs and contains up to 12 copies each of the two minor structural proteins, the RNA-directed RNA polymerase (RdRP) VP1 and the capping enzyme VP3. VP1 is tethered to the inner surface of VP2 (McClain et al., 2010). The location of VP3 is unresolved but has been proposed to exist in complex with VP1 (Figure 1.Rotavirus and Figure 2.Rotavirus).
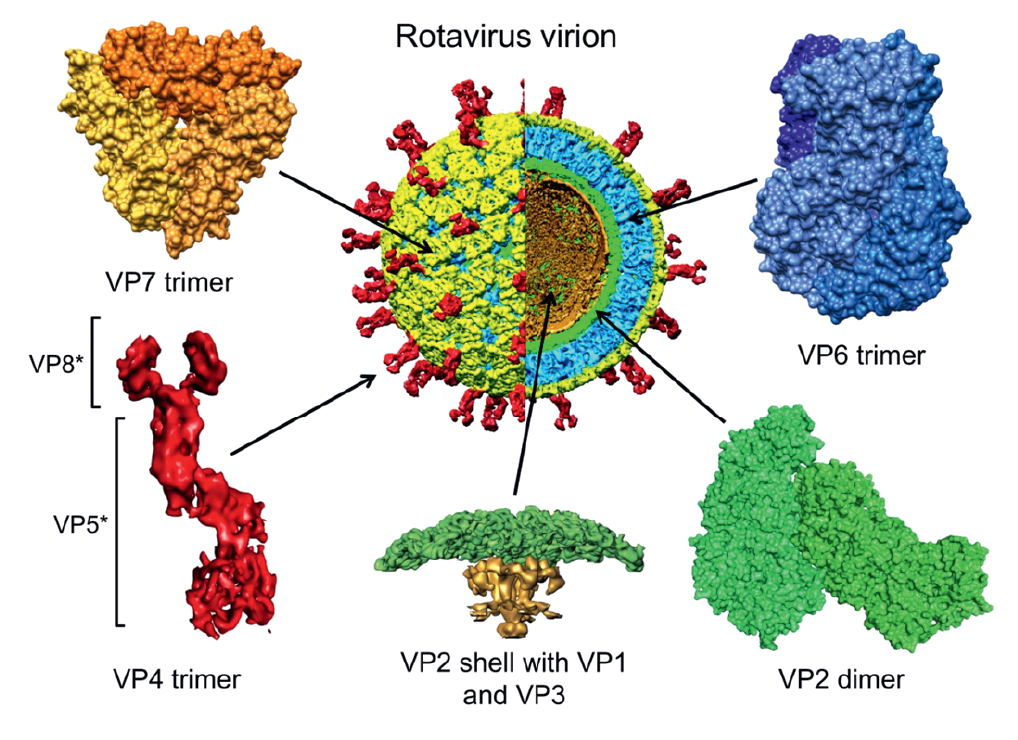 |
| Figure 2. Rotavirus Structure and location of protein components of a rotavirus virion. A cutaway view of a cryoEM image reconstruction of a rotavirion at 9.5 Å resolution (center) is a reference for enlarged, high-resolution images of specific virion components. The particle and components are colored as follows: VP4 spikes (red), VP7 layer (yellow), VP6 layer (blue) and VP2 layer (green). A portion of VP2 that extends into the interior of the core (the “hub”) and transcriptional enzymes VP1 and VP3 are colored gold in the enlarged image of a five-fold vertex (bottom, center). The VP8* and VP5* cleavage products of the VP4 spike are indicated. The trimeric foot and dimeric stalk and head of VP4 can be seen. PDB files 3KZ4 (VP6 and VP2 of Rotavirus A strain UK) and 3GZT (VP7 of rotavirus A strain RRV) were used to make images. (Courtesy of B. V. V. Prasad.) |
The assembled genomic dsRNAs, the RdRP and capping enzyme, and the VP2 layer, comprise the rotavirus core, which has a diameter of about 51 nm. Each of the two outer layers of the rotavirus virion is organized with T=13 icosahedral symmetry (Settembre et al., 2011). Spanning these layers is a characteristic set of 132 large channels that link the outer surface with the inner VP2 protein layer. The intermediate capsid layer is composed of 780 copies of VP6, arranged as 260 trimeric morphological units positioned at the local and strict three-fold axes of the icosahedral lattice (Figure 2.Rotavirus). The VP6 layer forms the outer surface of the double-layered particle (DLP; about 70.5 nm in diameter) and is directly comparable to one of the capsid layers of virions of some other genera within the order Reovirales (e.g., the VP7 (T13) layer of the orbiviruses). Two proteins (VP4 and VP7) form the outermost layer of the rotavirus virion (about 75 nm in diameter, not including spikes) and are required for infectivity (Figure 2.Rotavirus). The glycoprotein VP7 makes up the surface of the outermost shell, which is arranged as 260 trimers stabilized by Ca2+ bound in the inter-subunit interfaces. VP7 trimers cap the trimeric pillars of VP6 and grip them with amino-terminal arms. Projecting from the VP7 layer are 60 trimeric spikes formed by VP4. Trypsin cleavage of VP4 generates amino-terminal (VP8*) and carboxyl-terminal (VP5*) fragments of VP4 and primes virions for infectivity. Primed VP4 spikes are about 20 nm long and extend about 12 nm from the surface of the outer VP7 layer, giving a final maximum particle diameter of about 100 nm. The foot of the VP4 spike has three-fold symmetry and interacts extensively with VP6. Distal to the foot, the spike lacks three-fold symmetry. A stalk formed primarily by VP5* connects the foot to a paired body, atop which two heads formed by the VP8* receptor-binding domain sit. Each VP5* monomer contains a β-barrel domain, tipped by a hydrophobic apex, which is buried in the cleaved spike. Similar to enveloped virus fusion proteins, during entry VP4 undergoes a rearrangement that is linked to membrane penetration. The hydrophobic apices of VP5* are exposed as the β-barrel domains fold back and a trimeric coiled-coil zips up.
Physicochemical and physical properties
The rotavirus virion has a density of 1.36 g/cm3 in CsCl and sediments at 520–530S in sucrose. Virus infectivity is dependent upon the presence of the VP4-VP7 outermost protein layer, the integrity of which requires Ca2+. Treatment of virions with Ca2+-chelating agents, such as EGTA or EDTA, destabilizes VP7 trimers, leading to loss of the outer capsid. Infectivity is not affected by exposure of virions to pH ranges from 3 to 9 or, in the presence of 1.5 mM CaCl2, by storage for months at 4 °C or 20 °C. Infectivity is also relatively thermostable at 50 °C but can be lost by repeated cycles of freezing and thawing. Infectivity is generally resistant to fluorocarbon extraction, treatment with solvents such as ether and chloroform, or non-ionic detergents such as deoxycholate, all of which reflect the absence of a lipid-containing envelope on the mature particle. However, infectivity is lost by treatment with sodium dodecyl sulfate (0.1%), or a number of disinfectants such as betapropiolactone, chlorine, formalin and phenols. In addition, 95% ethanol, which disrupts the outer protein layer of the virion, represents an effective disinfectant. The VP4-mediated hemagglutinin activity of the infectious virion is lost rapidly at 45 °C or as a result of freezing and thawing. Some variation has been observed in the physicochemical properties and stability of intact virions of different rotavirus strains. For example, not all human rotaviruses show hemagglutinin activity, and they tend to lose the proteins of their outer layer more easily than some animal strains. In reassortment studies, some of this variation has been attributed to the parental origin of the VP4 component in the virion.
Double-layered particles (DLPs) are non-infectious, have a density of 1.38 g/cm3 in CsCl and sediment at 380–400S in sucrose. These are equivalent to cores of the orbiviruses. Single-layered rotavirus particles (cores) can be produced by treatment of DLPs with either chaotropic agents such as sodium thiocyanate or high concentrations of CaCl2. Cores have a density of 1.44 g cm−3 in CsCl, sediment at 280S in sucrose, and are readily disrupted by incubation in hypotonic solutions, and are structurally equivalent to sub-cores of the orbiviruses.
Nucleic acid
The rotavirus genome consists of 11 discrete segments of linear dsRNA, which are organized with dodecahedral symmetry in the core. For RVA-SA11, the segments range from 3,302 to 667 bp, giving a total of 18,555 bp. When resolved by gel electrophoresis, the segments of viruses in the species Rotavirus A (not including isolates from birds) typically display a 4:2:3:2 distribution pattern. The genome segments are numbered segment-1 (Seg1) to segment-11 (Seg11) in order of increasing mobility during electrophoresis, although the migration order of cognate segments, particularly for the Seg7 to Seg9 triplet, does vary. In some isolates of viruses of the species Rotavirus A, deviation from the 4:2:3:2 RNA migration pattern is indicative of a concatemerization event, in which up to an additional 1800 bp is packaged into viable virions, due to the partial duplication of a genome segment (Bányai et al., 2009). Rotavirus genomic RNA sequences are A+U rich (58–67%). The segments are completely base-paired, and the positive-sense strand contains a 5′-terminal cap structure (m7GpppG(m)GC), but lacks a polyadenylation signal near its 3′-end. In contrast to members of the genus Orthoreovirus, there is no evidence for the presence of single-stranded oligonucleotides within the virion. Two levels of terminal sequence conservation are evident for members of the species Rotavirus A. Firstly, all genome segments share short, conserved 5′- and 3′-termini. The 5′-terminal conserved region has a consensus sequence of 5′-(GGCUUUUAAA…)-3′, and the 3′-terminus has the consensus sequence 5′-(…AUGUGACC)-3′ (Table 1.Rotavirus) (Li et al., 2010). Immediately internal to these terminal regions at each end of the different segments, there is a second region of conservation of at least 30–40 bp, which is segment-specific. These two levels of sequence conservation may be indicative of cis-acting signals that are important for controlling transcription, replication, assortment, and gene expression. The 5′-nontranslated regions (NTRs) vary in length, but are typically less than 50 bp and are followed by at least one long ORF beginning with the first AUG. Some segments contain additional in-frame (Seg7, Seg9 and Seg10) or out-of-frame (Seg11) ORFs. However, only in the case of Seg9 and Seg11 are alternate start codons used to initiate synthesis of more than a single primary translational product from each segment. The 3′-NTRs vary in length from 17 bp (Seg1) to 182 bp (Seg10).
Compared to members of rotavirus A, detailed genomic information for members of other rotavirus species is sparse. However, complete sequence information is available for all genome segments of at least one isolate of one virus for each species. In all cases, the genome is made up of 11 segments with broadly similar properties to RVA in terms of length, presence of ORFs and conserved terminal sequences. However, in some rotavirus groups, additional ORFs have been identified (e.g., two ORFs encode NSP1-1 and NSP1-2 in Seg5 in members of Rotavirus B, Rotavirus G and Rotavirus I; and two ORFs in segments encoding VP6 and NSP5 of Rotavirus J).
Table 1.Rotavirus Conserved rotavirus terminal sequences (positive-sense strand)
|
Virus species |
Virus names |
5′-end |
3′-end |
|
Rotavirus A |
rotavirus A (RVA) |
5′-GGC(U/A)2U(A/U)4 |
(A/U)U(G/A)UG(A/G)CC-3′* |
|
Rotavirus B |
rotavirus B (RVB) |
5′-GG(U/C)(A/U)N(A/U)5** |
(A/U)3(A/G)2A(C/A)CC-3′ |
|
Rotavirus C |
rotavirus C (RVC) |
5′-GCC(A/U)7 |
UGUGGCU-3′ |
|
Rotavirus D |
rotavirus D (RVD) |
5′-GG(U/C)(A/U)4AA(A/U) |
(U/A/C)U(G/A/U)(U/C)GACC-3′ |
|
Rotavirus F |
rotavirus F (RVF) |
5′-GGC(A/U)3U(A/U) 3 |
UN(U/A/C)GACC-3′ |
|
Rotavirus G |
rotavirus G (RVG) |
5′-GG(A/C)A(A/U/G)6 |
(A/U)3AAA(G/A)ACCC-3′ |
|
Rotavirus H |
rotavirus H (RVH) |
5′-GG(C/A/U)A(C/A/U) |
(A/G)UA(U/C)ACCC-3′ |
|
Rotavirus I |
rotavirus I (RVI) |
5′-GG(U/C)A |
A5CCC-3′ |
|
Rotavirus J |
rotavirus J (RVJ) |
5′-GG(A/C)A |
A5 TA(T/C)ACCC-3′ |
* For Seg5, the 3′-terminal sequence is CUGUGAACC-3′.
** Rarely, a G or C is found at position 6, 8, or 10.
Proteins
Thirteen primary gene products have been defined for RVA-SA11. The majority of genome segments encode a single protein, but two segments (Seg9 and Seg11) each encode two primary translation products. In the case of Seg9, two initiation codons in the same reading frame may be used, giving largely overlapping forms of the protein product VP7. Gene 11 contains two out-of-frame ORFs, translation of which results in two unrelated non-structural proteins, NSP5 and NSP6 (Table 2.Rotavirus).
Table 2.Rotavirus Genome segments and protein products of RVA-SA11
|
Genome segment |
bp |
Protein |
Protein mass (kDa)* |
Protein copy number/particle |
Location |
Function |
|
Seg1 |
3302 |
VP1 (Pol) |
125.0 |
≤12 |
core |
RdRP; minor core component activated by VP2 |
|
Seg2 |
2690 |
VP2 (T1) |
102.4 |
120 |
innermost capsid |
Core shell protein with RNA binding activity; sub-group specificity antigen. T=1 symmetry with two molecules in the icosahedral asymmetric unit (interpreted in the orbiviruses as T=2 symmetry); |
|
Seg3 |
2591 |
VP3 (Cap) |
98.1 |
≤12 |
core |
Capping enzyme. Minor core component with guanylyltransferase, methyltransferase, 2'-5' oligoadenylate phosphodiesterase and ssRNA binding activities |
|
Seg4 |
2362 |
VP4 |
86.8 |
180 |
outer capsid |
Homotrimeic, viral attachment spike protein activated by trypsin cleavage to generate VP5* (membrane penetration) and VP8* (carbohydrate binding, with haemaglutination activity) moieties. P-type neutralization antigen. |
|
|
|
VP5* |
60.0 |
|
|
|
|
|
|
VP8* |
28.0 |
|
|
|
|
Seg5 |
1611 |
NSP1 |
58.7 |
0 |
cytoplasm |
Antagonist of interferon expression. Putative viral E3 ubiquitin ligase, with RNA binding activity and a RING domain |
|
Seg6 |
1356 |
VP6 (T13) |
44.8 |
780 |
intermediate capsid |
Trimeric major inner virus protein with T=13 symmetry. Group and sub-group specificity antigen. |
|
Seg7 |
1104 |
NSP3 |
34. 6 |
0 |
cytoplasm |
Binds 3′-end of viral mRNA and cellular eIF4G; promotes circularization of viral mRNAs. Surrogate of PABP, inhibits host translation |
|
Seg8 |
1059 |
NSP2 (ViP) |
36.7 |
0 |
viroplasm |
Viral inclusion body or viroplasm matrix protein. Octamer with NTPase, RTPase, ssRNA binding and helix destabilizing activities. Essential viroplasm component that interacts with NSP5 |
|
Seg9 |
1062 |
VP7 (1) |
37.4 |
780 |
outer capsid |
Virion surface glycoprotein, forming Ca2+-stabilized trimer. G-type neutralization antigen, |
|
|
|
VP7 (2) cleaved form |
33.9 |
|
|
|
|
Seg10 |
751 |
NSP4 |
20.3 |
0 |
RER membrane |
RER transmembrane glycoprotein, binds DLPs, essential for budding into ER and addition of outer capsid, age-dependent diarrhea-inducing enterotoxin, disrupts Ca2+ homeostasis |
|
Seg11 |
667 |
NSP5 |
21.7 |
0 |
viroplasm |
Phosphorylated, O-linked glycosylated, and serine-threonine rich protein with RNA binding activity. Essential viroplasm component that interacts with NSP2 |
|
|
|
NSP6 |
11.0 |
0 |
viroplasm |
Product of second Seg11 ORF with RNA binding activity. Iinteracts with NSP5; non-essential viroplasm component |
* Viruses from other species within the genus may have proteins with significant differences in masses.
The nomenclature system employed for rotavirus proteins numbers them according to their migration rates upon SDS-PAGE, starting with the slowest (i.e., highest molecular weight). Structural proteins are given the prefix “VP”, whereas non-structural proteins are given the prefix “NSP”. Six structural proteins have been identified, and their approximate locations within the virion have been defined. The viral core, which encapsidates the dsRNA genome, is composed of three proteins. Two of the core proteins (VP1 and VP3) are directly associated with the genome, while the third (VP2) makes up the core shell, the innermost protein layer of the capsid. VP1, the largest viral protein (125 kDa), is a four-tunneled RdRP, responsible for both transcription and replication. Transcriptional activity can be detected in preparations of purified DLPs. Genome replication (negative-strand synthesis) can be achieved in vitro using disrupted viral core preparations or recombinant VP1 protein, in combination with VP2, which is necessary for its activity. Viral core preparations also have guanylyltransferase and methyltransferase activities. The VP3 component (98 kDa; the capping enzyme) of DLPs, binds ssRNA and forms adducts with GTP, S-adenosyl-l-methionine, and is an antagonist of the cellular antiviral 2′-5′ oligoadenylate synthetase-RNase L pathway (Ogden et al., 2015). VP1, possibly in complex with VP3, is tethered near the inward protruding hubs at the five-fold vertices of the VP2 capsid layer (Figure 2.Rotavirus). The amount of VP1 and VP3 in the virion is known to be low (≤12 molecules/particle) but has not been measured precisely. VP2 (102 kDa) is the most abundant protein of the viral core, with 120 molecules per virion. VP2 is required for encapsidation of VP1 and VP3 and has nonspecific ssRNA- and dsRNA-binding activity. By analogy with other reoviruses, it is likely that VP2 determines both the size of the particle as well as the structural organization of the outer capsid components, the internal enzymatic components and the RNAs. This important functional load is reflected in the highly conserved nature of VP2.
The intermediate protein layer of the virion is made up of 260 trimers of VP6 (45 kDa). The two remaining structural proteins of the virion, VP4 (87 kDa) and VP7 (36 kDa), of which there are 60 trimers and 260 trimers per virion, respectively, make up the outermost protein layer (Figure 2.Rotavirus). The spike protein VP4 contains a trypsin cleavage site approximately one-third of the way along its length. Cleavage of the protein in vitro by treatment with trypsin produces two products, VP5* (60 kDa) and VP8* (28 kDa), enhances virus infectivity, and induces conformational changes that stabilize the spike structure. The VP8* cleavage product has hemagglutinin activity and contains a carbohydrate binding site (galectin-like fold). VP7 is the primary component of the outer shell of the virion. The VP7 glycoprotein is synthesized on the rough ER (RER) and co-translationally inserted in the ER membrane. Seg9 contains two in-frame initiation codons, both of which may be used to generate VP7 protein products. However, post-translational cleavage results in removal of the amino terminus of both forms of VP7, yielding identical protein products that are incorporated into the virion.
Six non-structural proteins are encoded by the viral genome. NSP1 (59 kDa), encoded by Seg5, is the most variable of all the rotavirus proteins within members of a particular species; 65% amino acid sequence diversity is observed between viruses of Rotavirus A. The total length of this genome segment, as well as that of the ORFs, can vary among viruses in this genus. Although highly variable overall, NSP1 has a conserved cysteine-rich motif near the amino terminus, which is organized in a manner characteristic of zinc-binding RING-domain proteins. NSP1 has been shown to bind both the 5′-end of viral RNA and zinc. As indicated by studies of rotavirus reassortants in a mouse model of infection, NSP1 has a role in viral pathogenesis and is a virulence determinant. NSP1 antagonizes the host innate immune response by inducing the degradation of cellular proteins required for expression of type I interferon [interferon regulatory factors (IRF3, IRF5 and IRF7) and b-transducin repeat-containing protein (b-TrCP)], possibly by hijacking cellular E3 ubiquitin ligases. NSP1 may also play a role in host range restriction, suggesting that its activity may vary depending on the host species.
NSP2 (35 kDa) contains a histidine triad (HIT) nucleotide-binding domain and self-assembles into a stable octamer, the functional form of the protein. It binds nonspecifically to ssRNA, has NTPase, RTPase and helix destabilizing activities, and interacts with NSP5 to form viroplasms (equivalent to VIBs generated by members of other genera within the order Reovirales). Viruses that contain temperature-sensitive mutations in Seg8, which encodes NSP2, have an RNA-negative phenotype at the non-permissive temperature, indicating that NSP2 has a direct role in the mechanism of virus replication. It has been hypothesized that NSP2 functions as a molecular motor and plays an important role in viral RNA packaging.
NSP3 (34 kDa) is a multifunctional protein that forms dimers and has several binding partners. NSP3 binds specifically to the 3′-terminal conserved sequence of viral mRNAs via its amino terminus. The carboxyl-terminal region of NSP3 interacts with the eukaryotic translation initiation factor eIF4G, and its middle region binds RoXaN (rotavirus X protein associated with NSP3). In uninfected cells, the eukaryotic translation initiation factor eIF4G1 interacts with poly(A)-binding protein (PABP), which binds to the 3′-poly(A) tail of mRNAs and stimulates translation by circularizing host mRNAs. NSP3 is proposed to inhibit host cellular protein synthesis during infection by evicting PABP from its binding site on eIF4G. NSP3 may facilitate circularization of viral mRNA through its specific interactions with eIF4G and the 3′-conserved sequence of viral mRNAs, thereby promoting viral translation.
NSP4 (20 and 28 kDa) is synthesized on the RER as a transmembrane protein and may be post-translationally glycosylated. Both glycosylated and nonglycosylated forms of NSP4 are detected in infected cells. During later stages of virion maturation, membrane-associated NSP4 functions as a receptor for DLPs, aids in acquisition of VP4 and VP7 and budding through the ER membrane. NSP4 also functions as a viral enterotoxin that leads to Ca2+ release from internal stores in the ER and induction of age-dependent diarrhea in mice. An NSP4 cleavage product is secreted from infected cells and binds to integrins α1β1 or α2β1, triggering a signalling pathway that activates phospholipase C and elevates inositol 1,4,5-triphosphate, leading to Ca2+ release. This pathway is distinct from that of intracellularly expressed NSP4.
The two remaining non-structural proteins, NSP5 (26 kDa) and NSP6 (12 kDa), are encoded by two ORFs in the same viral gene. NSP5, which is serine-threonine rich, is post-translationally modified by both phosphorylation and glycosylation. The protein has ssRNA- and dsRNA-binding activities, forms dimers, and is essential for viroplasm formation and genome replication. Multiple phosphorylated isomers of NSP5 exist in the infected cell. Phosphorylation is stimulated by NSP2 in vivo but mediated by cellular kinases and possibly an NSP5 autokinase activity. Highly phosphorylated forms of NSP5 localize to viroplasms; the significance of phosphorylation to NSP5 function is unknown. Interactions of NSP2 and NSP5 are responsible for viroplasm formation and localization of core proteins. Interactions of NSP2 and NSP5 with core proteins regulate progeny core formation. A carboxyl-terminal domain of NSP5 is required for multimerization, hyperphosphorylation and interactions with NSP6. A potential ORF encoding NSP6 is conserved among many, but not all, virus isolates. However, NSP6 protein expression has been demonstrated for only a few isolates, and its function is not clearly defined. For RVA-SA11, NSP6 (11 kDa) is phosphorylated and localizes to viroplasms, where it may play a regulatory role in the self-association of NSP5.
Information on proteins encoded by viruses of species other than Rotavirus A is less abundant and primarily drawn from sequence analysis of virus genes. The majority of rotaviruses are predicted to encode a single protein product per segment. It is clear that members of other rotavirus species have homologs of the proteins characterized for Rotavirus A viruses. However, high levels of variation are observed between protein homologs (or putative protein products) of viruses of different species (e.g., >84% divergence for VP3 and >87% divergence for VP4), in comparison to the variation observed among Rotavirus A proteins (e.g., <25% divergence for VP2 and <45% divergence for VP4) (Figure 3.Rotavirus). In a few cases, proteins of viruses from species other than Rotavirus A have been analyzed. For example, the structure of NSP2 from a Rotavirus C virus has been determined. Although structurally quite similar, this protein was unable to complement the function of NSP2 in cells infected with the Rotavirus A virus RVA-SA11 in which expression of the cognate NSP2 had been knocked down. Additionally, it has been shown that recombinant VP1 from a Rotavirus C virus will replicate RNA from a Rotavirus A virus, or its own RNA, but only in the presence of its cognate VP2. The VP1 of a Rotavirus C virus is also predicted to have a structure similar to RVA-SA11 VP1. Unlike the NSP3 segment of Rotavirus A viruses, the analogous segment of Rotavirus C viruses codes for two proteins (NSP3 and dsRBP) through the activity of 2A translational element (Langland et al., 1994, Donnelly et al., 2001). Viruses in the species Rotavirus B, Rotavirus G and Rotavirus I encode two proteins – NSP1-1 and NSP1-2 – from Seg5 whose functions are unknown. Further insights into the structure and function of proteins encoded by viruses from species other than Rotavirus A await the establishment of suitable highly permissive cell culture systems and the expression and characterization of recombinant proteins from viruses other than those belonging to Rotavirus A.
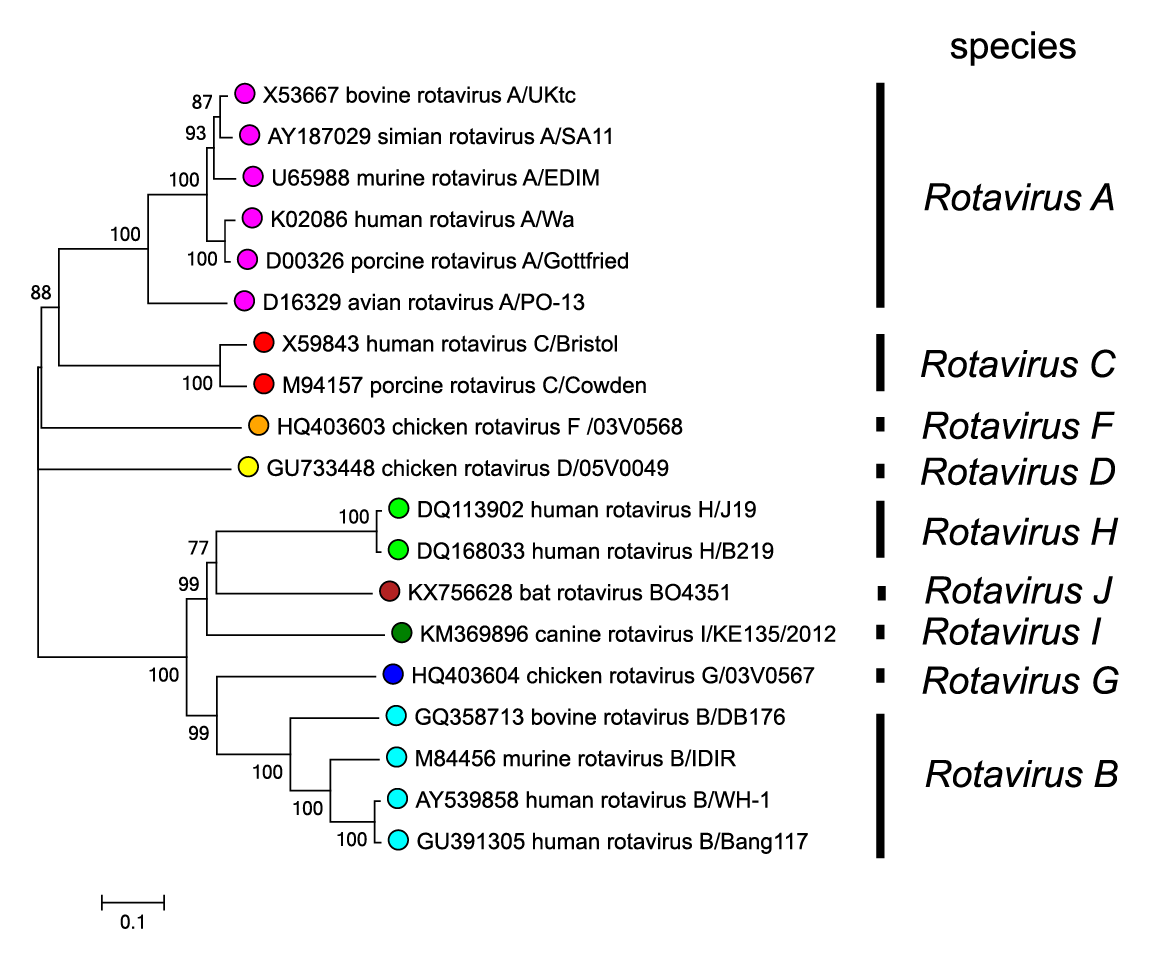 |
| Figure 3. Rotavirus Phylogenetic relationships between members of different rotavirus species using the amino acid sequences of the VP6 inner capsid protein. Sequences were aligned using MUSCLE (Edgar 2004) and the tree constructed in MEGA7 (Kumar et al., 2016) using the neighbour-joining method and the p-distance algorithm with pairwise deletion. Bootstrapping values (1000 replicates) above 70 % are shown. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Lipids
None reported. Immature particles acquire a transient membrane during budding into the ER.
Carbohydrates
Three viral proteins have been shown to be glycosylated. In two cases (VP7 and NSP4), the sugar is N-linked to asparagine, and in the third (NSP5) it is O-linked to serine and/or threonine.
Genome organization and replication
RNA-protein coding assignments have been fully described for several Rotavirus A viruses including RVA-SA11 (Table 2.Rotavirus). The replication cycle for many animal rotaviruses, which is typically completed in 12–15 h at 37 °C, has been studied primarily in continuous cell cultures derived from monkey kidneys. There is little definitive information about the early steps in the replication cycle. VP4 is the viral attachment protein but the cellular receptor has not been conclusively identified. Some rotavirus strains attach to N-acetyl-neuraminic (sialic) acid residues on the cell surface, and there is evidence to suggest that several integrins and heat shock protein 70 serve as co-receptors. Current data suggest that rotavirions may enter cells either by receptor-mediated endocytosis or by direct membrane penetration. In both cases, virion entry leads to loss of the outer VP4 and VP7 protein layer and release of a transcriptionally-active DLP into the cytoplasm. DLP-associated enzymes produce 5′-capped, non-polyadenylated mRNAs, which are synthesized using the full-length negative-sense strand of each genome segment as a template. In part, gene expression is regulated by differences in the level of transcription occurring from each genome segment. The virus mRNAs serve two functions. Firstly, they are used for translation, producing virus proteins at levels that are regulated by the efficiency of mRNA translation. Secondly, viral mRNAs act as templates for negative-sense strand synthesis, producing dsRNA genome segments. The use of short interfering RNA technology has provided evidence that the pools of mRNAs that serve as templates for translation and replication are distinct. Virion assembly begins in cytoplasmic inclusions termed viroplasms, formation of which is mediated by NSP2 and NSP5. During early assembly steps, the 11 different viral mRNAs interact with one another and with VP1 and VP3. These interactions are followed by association with VP2, which triggers negative-sense strand synthesis and results in the formation of cores containing a complete set of 11 dsRNA segments. Cis-acting replication elements for negative-sense strand synthesis have been identified using an in vitro replication system; most critical are the seven 3′-terminal nucleotides of the mRNAs. Following the assembly of cores, VP6 is added, forming DLPs. The next steps in the morphogenesis of progeny virions are unique to rotaviruses and involve recruitment of DLPs to the ER by the transmembrane glycoprotein NSP4. Budding of DLPs through the ER results in the transient acquisition of a lipid envelope and addition of VP4 and VP7 to form the outer virion shell. Newly formed TLPs can be released by either cell lysis or exocytosis.
Biology
Rotaviruses can be difficult to cultivate in vitro, with highly permissive growth generally restricted to epithelial cell lines derived from monkey kidneys. Infection can be enhanced by pre-treatment of virions with trypsin. The restriction of virus growth in vitro parallels the in vivo situation, in which virus replication is typically restricted to the terminally-differentiated enterocytes lining the tips of the microvilli in the small intestine. However, evidence suggests that some rotavirus infections may cause antigenemia and viremia due to replication at secondary sites in the host. This may occasionally lead to extraintestinal disease manifestations, e.g. encephalopathy, which is described in a low percentage of rotavirus infections (Goldwater et al., 2001, Kawamura et al., 2014).
There are several mechanisms of pathogenesis, including the destruction of enterocytes, leading to malabsorption and an osmotic diarrhea. Prior to the appearance of histologic changes, a watery diarrhea is often seen, which is thought to be secretory, possibly induced by the action of the rotavirus enterotoxin NSP4. It has been proposed that the destruction of enterocytes causes a loss of the permeability barrier between the gut lumen and the vasculature, resulting in the osmotic pull of fluid from the circulation into the gut and the watery characteristic of rotavirus-induced diarrhea. Rotaviruses infect a wide range of avian and mammalian species, with disease being restricted in the great majority of cases to the young. Rotavirus A, Rotavirus B, Rotavirus C and Rotavirus H viruses can infect humans, with Rotavirus A viruses being responsible for the majority of seasonal endemic diarrheal disease in young children. Members of Rotavirus A can also infect other mammals (e.g., pig, cattle, horse, rat, mouse and bat) in which they cause diarrhea, and birds (e.g., chicken, turkey and pigeon) in which they cause enteric signs and runting and stunting syndrome. Rotavirus B viruses have caused sporadic epidemic outbreaks of gastroenteritis in adults, but pigs are their main reservoir (Marthaler et al., 2014, Joshi et al., 2019). Rotavirus C viruses have been associated with self-limiting outbreaks of gastroenteritis in humans, primarily in the young, but are also widely distributed in animals, mainly pigs (Marthaler et al., 2014, Trovão et al., 2019). Rotavirus D, Rotavirus F and Rotavirus G viruses have been exclusively identified in birds (Dhama et al., 2015). Rotavirus H viruses infect humans, pigs and bats (Yinda et al., 2018), Rotavirus I viruses have been detected in dogs, cats and sea lions (Mihalov-Kovács et al., 2015, Phan et al., 2017, Li et al., 2011), and Rotavirus J viruses have recently been described in bats (Bányai et al., 2017).The species Rotavirus E was created in 1999 based on genome profiling and comparative terminal fingerprinting (Pedley et al., 1986). However, due to the lack of any subsequent virus isolates or sequence data, it was removed as a species by the ICTV in 2019.
Antigenicity
Three viral proteins (VP4, VP6 and VP7) of Rotavirus A viruses have been subjected to detailed antigenic characterization. VP6, which forms the intermediate capsid layer, is a highly conserved and highly immunogenic protein, carrying species-specific determinants, previously also referred to as groups and subgroups (SG)-specific determinants. VP6 does not elicit the production of neutralizing antibodies, but it may play a role in the induction of protective immunity. VP6 has been the major target of diagnostic assays for rotaviruses and can be used to discrimintate between members of different species and unclassified viruses.
Among Rotavirus A viruses, VP6 bears epitopes that allow antigenic classification of rotavirus into SG I, SG II, both SG I and II, or neither SG, according to reactivity with two monoclonal antibodies. More than two decades ago, a binary classification system was established reminiscent of the one used for the classification of influenza A viruses based on the antigenicity of outer capsid proteins. This system was based on the antigenic reactivities of the two outer capsid proteins, VP4 and VP7, which independently elicit neutralizing antibodies. Thus, rotavirus strains were classified into P serotypes (VP4 is protease sensitive) and G serotypes (VP7 is a glycoprotein). Classification of rotaviruses into P (VP4) or G (VP7) serotypes is performed by cross-neutralization assays using hyperimmune sera raised to prototype viruses or laboratory-engineered mono-reassortants. So far, 14 G serotypes and 14 P serotypes have been identified. As the ease of nucleic acid sequencing has increased, antigenic classification has slowly been replaced by the classification of rotaviruses into P (VP4) and G (VP7) genotypes that is based on nucleotide sequence identities of the VP4 and VP7 genes. G serotype designations largely coincide with G genotype designations, but there is greater discrepancy among P serotype and genotype designations. Thus, a dual nomenclature has been adopted for VP4 antigenic and genetic classification. The P serotype, when known, is denoted by a number. The P genotype is denoted by a number within squared brackets, which immediately follows the P serotype. When the P serotype is not known, only the P genotype is used.
A uniform classification and nomenclature system, based on nucleotide identities of the 11 rotavirus genome segments and phylogenetic dendrograms, has been established by the Rotavirus Classification Working Group for Rotavirus A (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg) (Matthijnssens et al., 2008, Matthijnssens et al., 2011). This classification scheme currently comprises 36 G (VP7), 51 P (VP4), 26 I (VP6; inner capsid), 22 R (VP1; RdRP), 20 C (VP2; core), 20 M (VP3; methyltransferase), 31 A (NSP1; interferon antagonist), 22 N (NSP2; NTPase), 22 T (NSP3; translation enhancer), 27 E (NSP4; enterotoxin), and 22 H (NSP5/6; phosphoprotein) genotypes. A similar classification system has been described for viruses of Rotavirus B (Shepherd et al., 2018) and Rotavirus C (Suzuki and Hasebe 2017).
In the case of viruses in species other than Rotavirus A and Rotavirus C, little information is available at present on the extent of their antigenic diversity.
Species demarcation criteria
Rotaviruses are currently grouped into nine species (Rotavirus A, Rotavirus B, Rotavirus C, Rotavirus D, Rotavirus F, Rotavirus G, Rotavirus H, Rotavirus I and Rotavirus J). Viruses within different species are thought to be unable to reassort their genome segments under normal circumstances, and each species may, therefore represent, a separate gene pool.
In addition to the other general criteria used throughout the family, members of a species in the genus Rotavirus may be identified by:
- Serological cross-reactivity by ELISA, using either polyclonal sera or monoclonal antibodies against VP6.
- Sequence analysis of conserved genome segments (e.g., Seg1 and Seg6). A cut-off value of 53% identity of the VP6 amino acid sequence has been used to differentiate members of different rotavirus species (Matthijnssens et al., 2012).
- Host range. For example, isolates of Rotavirus D, Rotavirus F and Rotavirus G have only been detected in avian species.

