Family: Phenuiviridae
Genus: Uukuvirus
Distinguishing features
Uukuviruses were originally isolated from Ixodes ticks in 1976 from Finland. Twenty-three species for 28 viruses are assigned to the genus Uukuvirus. The uukuvirus genome has four genes, encoding a large protein (L), external glycoproteins (Gn and Gc), a nucleocapsid protein (N) and a non-structural protein (NSs), this gene content/genome organisation being similar to that of bandaviruses and mobuviruses. Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences, viruses classified in the genus Uukuvirus form a monophyletic cluster clearly distinguished from other phenuivirids (Oker-Blom et al., 1964, Simons et al., 1990, Palacios et al., 2013).
Virion
Morphology
Virions have an enveloped structure and are spherical or pleomorphic, 90–100 nm in diameter with surface glycoprotein projections of 5–10 nm which are embedded in a lipid bilayer envelope approximately 5 nm thick.
Nucleic acid
The uukuvirus genome encompasses three single-stranded segments of negative-sense or ambisense RNA. The terminal nucleotides of each segment occur in a canonical, conserved sequence (in coding sense) 5′-ACACAAAGAC…CUCUUUGUGU-3′ and may form panhandle structures typical of other members of the class Bunyaviricetes (Table 2 Phenuiviridae). The L segment (6.3–6.4 kb) encodes a protein with a predicted molecular mass of 240–250 kDa that is homologous with the bunyaviral RNA-directed RNA polymerase (RdRP) domain. The M segment (3.3–3.4 kb) encodes a protein of 112–114 kD that is homologous with the phlebovirus glycoprotein G1 and G2 sequences, and also with the phlebovirus glycoprotein C-terminal Ig-like domain. The S segment (1.7–1.9 kb) encodes two proteins, one of 27–51 kDa that is homologous with the tenuivirus/phlebovirus N domain, and a non-structural protein of 11–35 kDa with unknown function (Table 3 Phenuiviridae) (Simons et al., 1990, Palacios et al., 2013).
Proteins
Mature virions contain three major structural proteins of approximately 57, 55 and 27–51 kDa that correspond to the Gn, Gc and N proteins of phenuivirids.
Genome organization and replication
Uukuvirus genome arrangement is similar to that of bandaviruses and mobuviruses (Figure 1 Uukuvirus). Many uukuvirus genomes encompass three single-stranded segments, but some lack the M segment and consist of two segments. The L, M, and S segments putatively encode L, a glycoprotein precursor (comprising Gn and Gc) and N, respectively. The Gn and Gc glycoproteins of 54–59 kDa and 55 kDa were earlier referred to as G1 and G2 based on apparent size following gel electrophoresis. The S segment exhibits an ambisense coding strategy; two ORFs, N and NSs, are separated by a noncoding intergenic region that potentially forms a long A/U rich stem-loop structure. In some two-segemented uukuviruses, the S segment encodes only N on the virus-complementary RNA. (Simons et al., 1990, Palacios et al., 2013). Replication, morphogenesis, assembly, and budding are described in ‘Genome organisation and replication’ section on the family page.
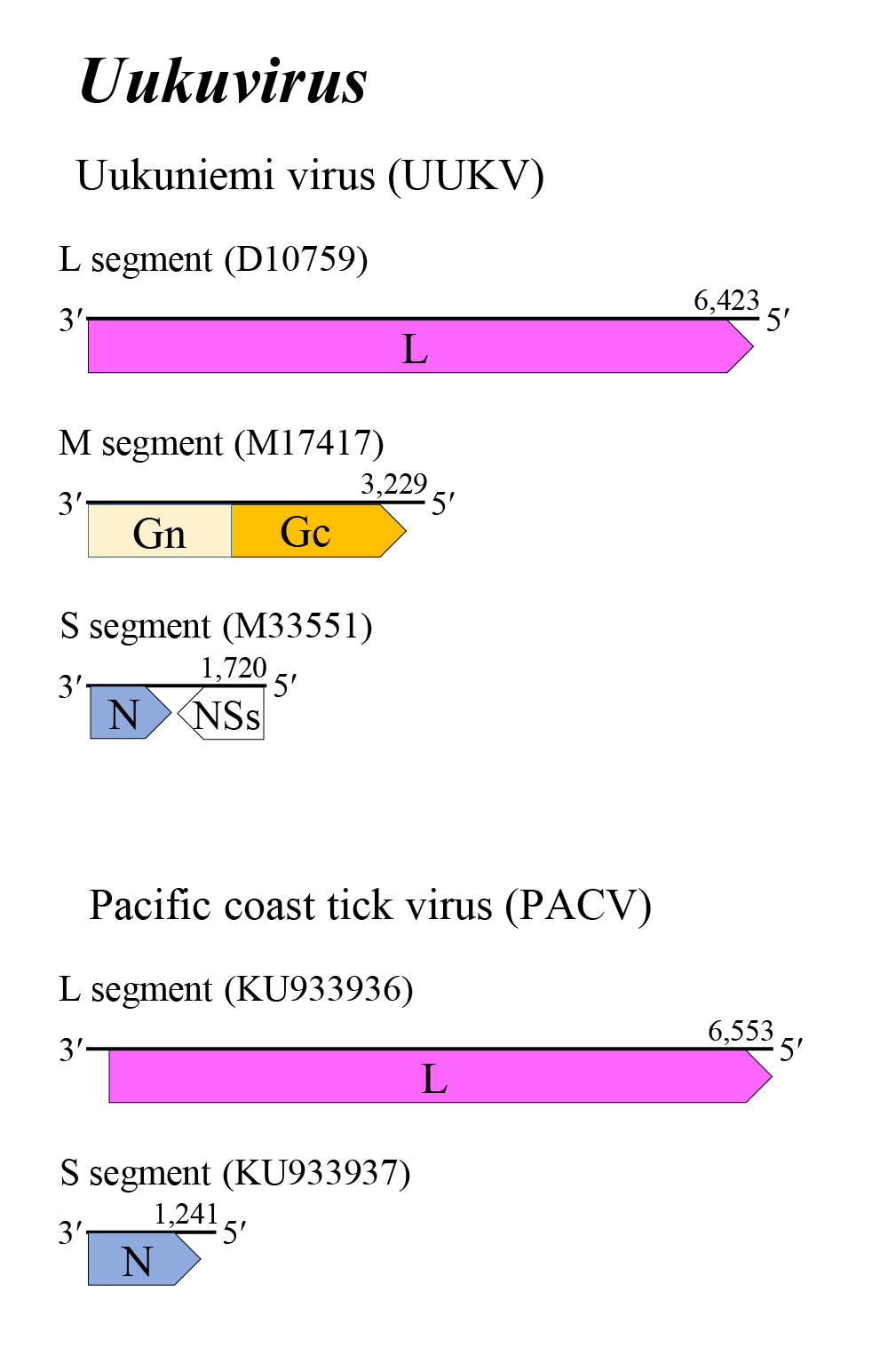 |
| Figure 1 Uukuvirus. Genome organization of some uukuviruses. Coloured boxes depict ORFs that encode N, nucleocapsid protein; Gn and Gc, external glycoproteins; and L, large protein. A white box depicts an ORF that encodes NSs, non-structural protein. |
Biology
Host range
Most uukuviruses infect mammals and birds and are transmitted by ticks. However, some uukuviruses replicate only in ticks. Hosts of uukuviruses include cattle, mice, rodents, reindeer, common murres [Uria aalge (Pontoppidan, 1763)] and humans. In general, uukuviruses have not been considered to be of public health and agricultural significance, although antibodies to some uukuviruses have been detected in sera from humans, cattle, wild animals, reptiles and birds (Hubálek and Rudolf 2012, Palacios et al., 2013).
Transmission
Uukuviruses are transmitted by particular species of ticks in a circulative, propagative manner. The major vectors are Argas robertsi (Hoogstraal, Kaiser & Kohls, 1968), A. reflexus (Fabricius, 1794), Dermacentor marginatus (Sulzer, 1776), D. nuttalli (Olenev, 1928), D. occidentalis (Marx, 1892), D. variabilis (Say, 1821), Haemaphysalis flava (Neumann, 1897), H. hystricis (Supino, 1897), H. leporispalustris (Packard, 1869), H. longicornis (Neumann, 1901), Ixodes ricinus (Linnaeus, 1758), I. uriae (White, 1852), Rhipicephalus microplus (Canestrini, 1888), and R. pulchellus (Gerstacker, 1873). Transovarial and venereal transmission have been demonstrated for some uukuviruses in their arthropod vectors. In addition to transmission by arthropod vectors, mammals can become infected through contact with the blood or body fluids of infected animals. Several uukuviruses have been isolated from the blood of common murres [Uria aalge (Pontoppidan, 1763)] and from ticks collected from nests of cliff swallows (Petrochelidon pyrrhonota Vieillot); avian host and/or vector movements may result in virus dissemination (Palacios et al., 2013, Li et al., 2015, Matsuno et al., 2015, Ejiri et al., 2018).
Species demarcation criteria
The criteria demarcating species in the genus are:
• Less than 95% identity in the amino acid sequence of the RdRP
Phylogenetic relationships across the genus have been established from maximum likelihood trees generated from full sequences of L proteins (Figure 2 Uukuvirus).
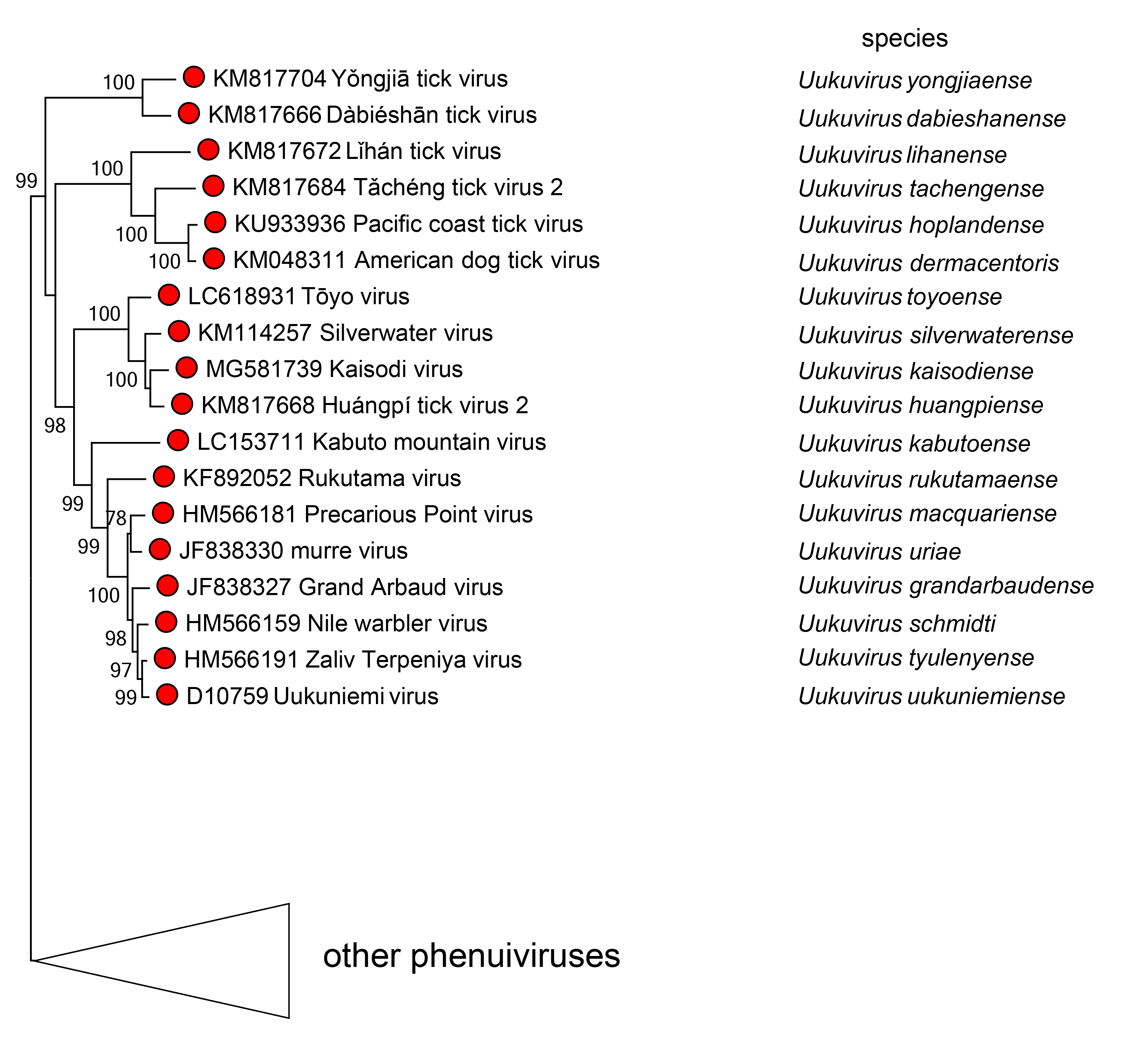 |
| Figure 2 Uukuvirus. Phylogenetic analysis of uukuvirus species. Phylogenetic reconstruction is based on a MAFFT-alignment of the RdRP amino acid sequences of phenuivirids using E-INS algorithm. The ML phylogenetic tree was inferred using RaxML-NG performing 1,000 bootstrap replicates. Trees were inferred under the WAG substitution model. Tree branches are proportional to genetic distances between sequences and the scale bars at the bottom indicate substitutions per amino acid. Full tree shown in Figure 3 Phenuiviridae. For alignment and treefile see Resources section. |
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Balambala tick virus | L: MW561967; S: MW561971 | BaTV |
| Bole tick virus 1 | L: KM817664; S: KM817731 | BTV1 |
| brown dog tick phlebovirus 1 | L: MN025506; S: MN025507 | BDTPV1 |
| brown dog tick phlebovirus 2 | L: OM326751; S: OM326753 | BDTPV2 |
| Catch-me-cave virus | L: OK493375; M: OK493376; S: EU274384 | CMCV |
| Chāngpíng tick virus 1 | L: KM817665; S: KM817732* | ChaTV |
| Cheeloo uukuvirus | L: OR115134; S: OR115135 | CUV |
| Dermacentor reticulatus uukuvirus | L: ON684362; S: ON684363 | DeRV |
| Dermacentor uukuvirus | L: OP863283; S: OP863282 | DeUV |
| Manawa virus | L: JQ924565; M: JQ924566; S: JQ924567 | MWAV |
| Qīngdǎo tick uukuvirus | L: OQ513654; M: OQ513655; S: OQ513656 | QiTV |
Virus names and virus abbreviations are not official ICTV designations.

