Family: Phenuiviridae
Genus: Coguvirus
Distinguishing features
The six viruses currently assigned to the genus Coguvirus were discovered in a wide range of dicotyledonous plants. Coguviruses have a genome consisting of a large negative-sense RNA (RNA1) and a small ambisense RNA (RNA2). These RNAs contain three genes, encoding two structural proteins, a large protein (L) and a nucleocapsid protein (N), and one putative non-structural protein likely encoding a movement protein (MP) involved in cell-to-cell movement in plant hosts. The apparent lack of a membrane-bound virus particle distinguishes coguviruses from other viruses in the family Phenuiviridae. Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences, viruses classified in the genus Coguvirus form a monophyletic cluster clearly distinguished from other phenuivirids (Navarro et al., 2018a, Navarro et al., 2018b).
Virion
Morphology
Virus particles are filamentous and flexuous, 200–300 nm long and 6 nm in diameter; no envelope has been observed (Navarro et al., 2018a).
Nucleic acid and Proteins
The coguvirus genome consists of either two RNA segments: RNA1 (6.7 kb) and RNA2 (2.7 kb). The terminal nucleotides of each segment occur in a canonical, conserved sequence (in coding sense) 5′-ACACAAAGAU… AUCUAUGUGU-3′ and may form panhandle structures similar to those of other members of the class Bunyaviricetes (Table 2 Phenuiviridae). In silico analysis of coguvirus putative ORF sequences suggests that the coguvirus genome encodes two proteins with predicted molecular masses of 250–252 kDa and 39–42 kDa, homologous with the bunyaviral RNA-directed RNA polymerase (RdRP) domain and the tenuivirus/phlebovirus N domain, respectively. The genome may also encode a third protein of 45–53 kDa that shares sequence homology and/or structural characteristics with the MP of plant viruses (Table 3 Phenuiviridae) (Navarro et al., 2018a, Navarro et al., 2018b).
Genome organization and replication
Coguviruses have two segments (Figure 1 Coguvirus). RNA1 encodes the L protein that includes the putative RdRP domain. RNA2 exhibits an ambisense coding strategy; the two ORFs of N and MP are separated by a noncoding intergenic region that potentially forms a long A/U rich stem-loop structure. Since most plant viruses encode viral suppression proteins that counteract plant antiviral defence mechanisms based on RNA silencing, one or more of the proteins encoded by coguviruses may interfere with RNA silencing. Details of virus replication are unknown (Navarro et al., 2018a, Navarro et al., 2018b, Kormelink et al., 2021).
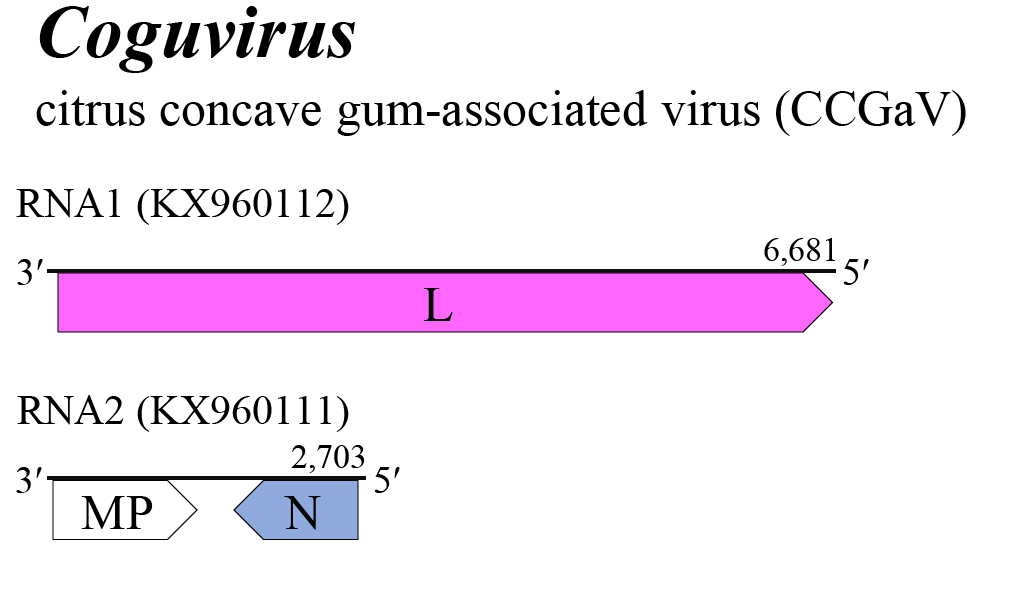 |
| Figure 1 Coguvirus. Genome organization of a coguvirus. Coloured boxes depict ORFs that encode N, nucleocapsid protein and L, large protein. White boxes depict ORFs that encode MP, non-structural cell-to-cell movement protein. |
Biology
Host range
Hosts of coguviruses are plants of the families Brassicaceae: Chinese cabbage [Brassica rapa subsp. chinensis ((L.) Hanelt, 1986)]; Cucurbitaceae: watermelon [Citrullus lanatus (Thunb.) Matsum & Nakai, 1916)]; Melanthiaceae: paris medicinal plants [Paris polyphylla (Smith, 1860)]; Rutaceae: sweet orange [Citrus sinensis ((L.) Osbeck, 1765)], grapefruit [C. paradisi (Macfad, 1830)], rough lemon [C. jambhiri (Lush, 1954)] and tangor [Citrus nobilis (Lour, 1931)]. (Xin et al., 2017, Navarro et al., 2018a).
Transmission
Coguviruses are transmitted by grafting. Mechanical transmission using sap extracts is possible with watermelon crinkle leaf-associated virus 1 and 2 (Navarro et al., 2018a, Navarro et al., 2018b, Xin et al., 2017).
Species demarcation criteria
The criteria demarcating species in the genus are:
• Less than 95% identity in the amino acid sequence of RdRP.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| blackberry line pattern virus | RNA1: ON624095; RNA2: ON624096 | BlaLPV |
| Edgeworthia chrysantha mosaic-associated virus | RNA1: ON602044; RNA2: ON602045 | ECMaV |
Virus names and virus abbreviations are not official ICTV designations.

