Family: Phenuiviridae
Genus: Mobuvirus
Distinguishing features
Four viruses, Browner virus (BRWV), Euproctis pseudoconspersa bunyavirus (EPBV), Mothra virus (MTHV) and Ñarangue virus (NRGV), are assigned to the genus Mobuvirus. The mobuvirus genome has four genes, encoding a large protein (L), a glycoprotein precursor (GP), a nucleocapsid protein (N) and a non-structural protein (NSs). Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences, viruses classified in the genus Mobuvirus form a monophyletic cluster clearly distinguished from other phenuivirids (Makhsous et al., 2017, Harvey et al., 2019).
Virion
Morphology
Virion morphology is unknown.
Nucleic acid and Protein
The mobuvirus genome encompasses three single-stranded segments of negative-sense RNA. Analysis of the terminal sequences has yet to be completed. The L segment (7.0–7.4 kb) encodes a protein with a predicted molecular mass of 256–273 kDa that is homologous with the bunyaviral RNA-directed RNA polymerase (RdRP) domain. The M segment (1.9–2.0 kb) encodes a protein of 64–68 kDa that is considered to be a glycoprotein precursor (GP). Unlike other phenuivirid GPs, the mobuvirus GP is not homologous with the conserved phlebovirus glycoproteins G1 and G2. The S segment (1.5 kb) of MTHV encodes a protein of 35–48 kDa that is homologous with the tenuivirus/phlebovirus N domain, and also a protein of 17 kDa of unknown function (Table 3 Phenuiviridae) (Makhsous et al., 2017, Harvey et al., 2019).
Genome organization and replication
Mobuvirus genome arrangement is similar to those of beidivirus, citriciviruses, goukoviruses, hudiviruses, hudoviruses, phasiviruses, pidchoviruses and tanzavirusess. The L and M segments putatively encode L and the glycoprotein precursor, respectively (Figure 1 Mobuvirus). The S segment of MTHV has two open reading frames (N and NSs), while the S segments of EPBV and NRGV encode only N on the virus-complementary RNA. Details of virus replication are unknown (Makhsous et al., 2017, Harvey et al., 2019).
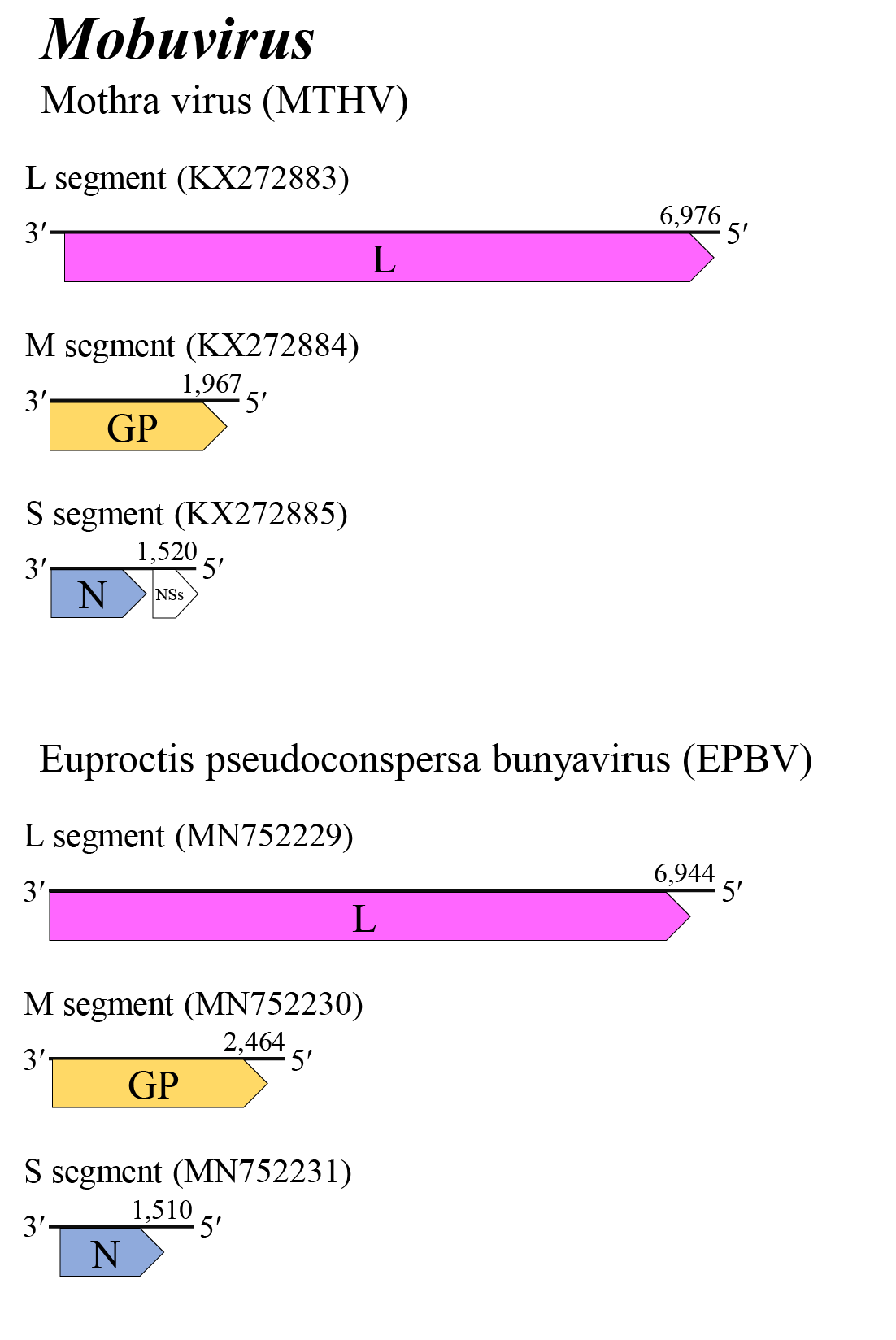 |
| Figure 1 Mobuvirus. Genome organization of mobuviruses. Coloured boxes depict ORFs that encode N, nucleocapsid protein; GP, glycoprotein precursor; and L, large protein. A white box depicts an ORF that encodes NSs, non-structural protein. |
Biology
BRWV was detected in a pooled extract of RNA from the fleas Stephanocircus pectinipes (Rothschild, 1915) and S. harrisoni (Traub & Dunnet, 1973) sampled in Australia. EPBV and MTHV were discovered in tea tussock moths [Euproctis pseudoconspersa (Strand, 1923)] in China and coding moths [Cydia pomonella (Linnaeus, 1758)] in USA, respectively. NRGV was detected in mosquitoes [Mansonia titillans (Walker, 1848)] in Germany (Makhsous et al., 2017, Harvey et al., 2019).
Species demarcation criteria
The criteria demarcating species in the genus are:
• Less than 95% identity in the amino acid sequence of the RdRP
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Etutanne virus 1 | L: LC642718* | ETTV1 |
| Etutanne virus 2 | L: LC642716* | ETTV2 |
| Etutanne virus 3 | L: LC642717* | ETTV3 |
* Sequences do not comprise the complete genome segment.
Virus names and virus abbreviations are not official ICTV designations.

