Family: Phenuiviridae
Genus: Mechlorovirus
Distinguishing features
Two viruses, melon chlorotic spot virus (MeCSV) and Ramu stunt virus (RmSV) are assigned to the genus Mechlorovirus. The mechlorovirus genome consists of six or eight segments and has eight or thirteen genes, these encoding two structural proteins, a large protein (L) and a nucleocapsid protein (N), and six or eleven non-structural proteins of unknown function. With eight genome segments, MeCSV has the most complex genome structure described so far for a member of the family Phenuiviridae. The apparent lack of a membrane-bound virus particle distinguishes mechloroviruses from some other viruses in the family Phenuiviridae. Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences, viruses classified in the genus Mechlorovirus form a monophyletic cluster clearly distinguished from other phenuivirids (Gaafar et al., 2019, Lecoq et al., 2019, Mollov et al., 2016).
Virion
Morphology
Virions are thin filaments 2–2.5 nm in diameter with lengths (200–3,000 nm) proportional to the size of the encapsidated segmented genomic RNA. The filamentous particles may appear to be spiral-shaped, branched or panhandled. No envelope has been observed (Gaafar et al., 2019).
Nucleic acid and Protein
The mechlorovirus genome encompasses six or eight segments of negative-sense or ambisense RNA (Figure 1 Mechlorovirus). The terminal nucleotides of each segment occur in a canonical, conserved sequence (in coding sense) 5′-ACACAAAGUC…GACUUUGUGU-3′ and may form the panhandle structures typical of other members of the class Bunyaviricetes (Table 2 Phenuiviridae). RNA1 (8.9–9.1 kb) is a negative-sense RNA that encodes a protein with a predicted molecular mass of 338–341 kDa that includes a region homologous with the bunyaviral RNA-directed RNA polymerase (RdRP) domain. RNA2 (1.7–1.8 kb), RNA3 (1.6 kb), RNA5 (1.5 kb), RNA7 (1.5 kb), and RNA8 (1.4 kb) are all ambisense RNAs, each encoding two non-structural proteins. RNA4 (1.6 kb) of MeCSV and RNA5 (1.4kb) of RmSV are negative-sense RNAs and encode N (33 kDa, homologous with the tenuivirus/phlebovirus N domain) (Table 3 Phenuiviridae) (Gaafar et al., 2019, Lecoq et al., 2019, Mollov et al., 2016).
Genome organization and replication
The mechlorovirus genome comprises six or eight RNA segments, three or four of which are negative-sense and two or five ambisense, collectively encoding two structural proteins, an RNA-directed RNA polymerase (RdRP) and N, and eleven non-structural proteins of unknown function (Figure 1 Mechlorovirus). Most plant viruses encode a viral movement protein (MP) to enable cell-to-cell movement in plant hosts and the viral suppressors of RNA silencing (VSR) that counteract plant antiviral defences. One or more of the mechlorovirus proteins may enable cell-to-cell movement in plant hosts and possibly interfere with RNA silencing. Details of virus replication are unknown (Lin et al., 2019, Kormelink et al., 2021).
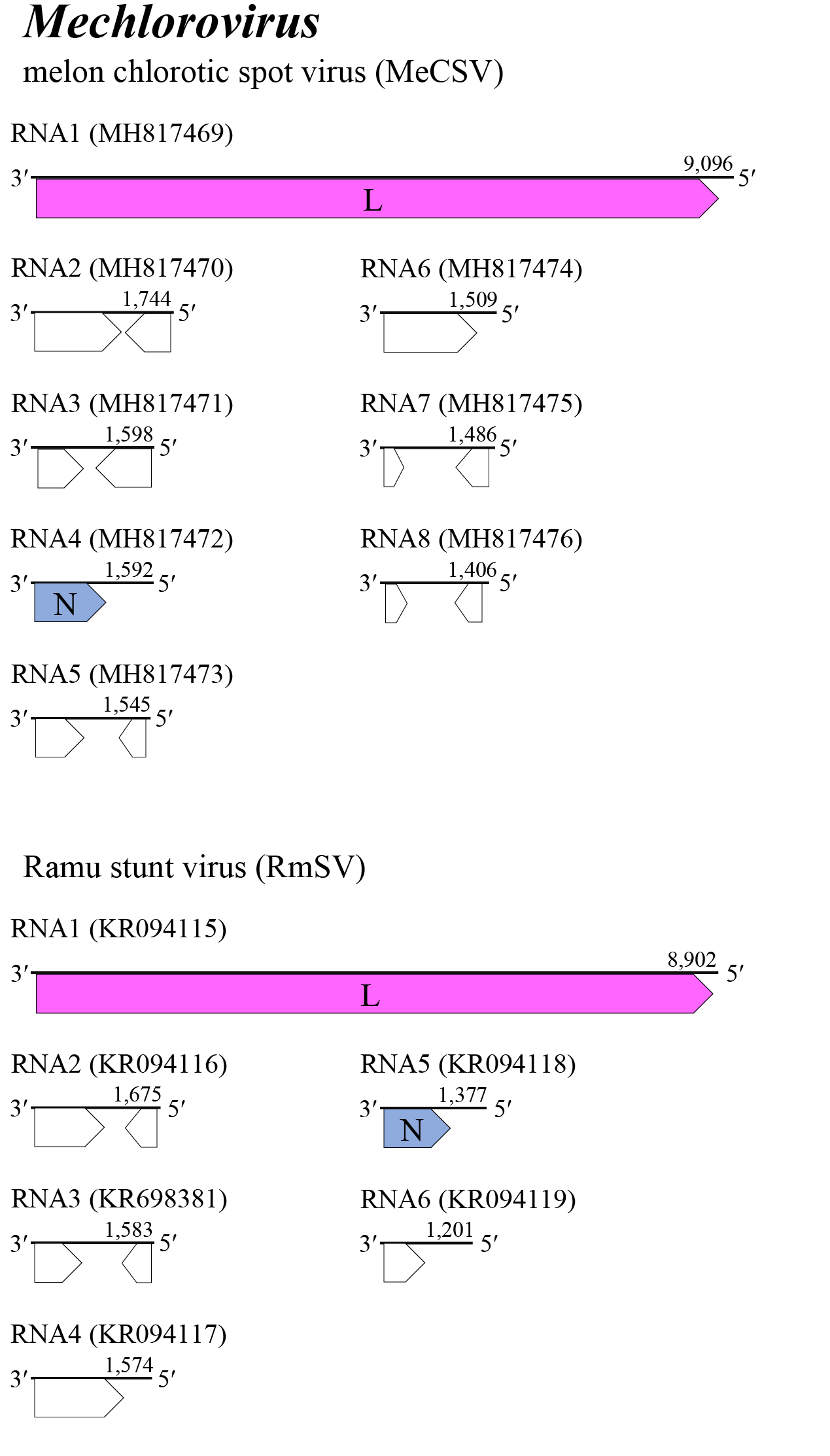 |
| Figure 1 Mechlorovirus. Genome organization of some mechloroviruses. Coloured boxes depict ORFs that encode N, nucleocapsid protein and L, large protein. White boxes depict ORFs that encode non-structural proteins of unknown function. |
Biology
Mechloroviruses have a relatively wide host range. MeCSV infects dicotyledonous plants of the families Chenopodiaceae: fat hen [Chenopodium amaranticolor (H.J.Coste & A.Reyn., 1907)], Cucurbitaceae: melon [Cucumis melo (L., 1753)], Fabaceae: alfalfa [M. sativa (L., 1753)], black medick [Medicago lupulina (L., 1753)], broad bean [Vicia faba (L., 1753)], and pea [Pisum sativum (L., 1753)], and Solanaceae: Nicotiana benthamiana (Domin, 1929), tobacco [N. tabacum (L., 1753)], Chinese lantern plant [Physalis pubescens (L., 1753)], petunia [Petunia x hybrida (hort. ex E.Vilm., 1863)], and tomato [Solanum lycopersicum (L., 1753)], and Polygonaceae: garden sorrel [Rumex acetosa (L., 1753)]. RmSV infests only Solanaceae: sugarcane [Saccharum officinarum (L., 1753)]. Ramu stunt disease caused by RmSV was first described in the mid-1980s associated with destructive problems in sugarcane production in Papua New Guinea. Mechanical transmission using sap extracts is possible with MeCSV. RmSV is transmitted by the planthopper Eumetopina flavipes (Muir, 1913), possibly in a circulative and propagative manner (Braithwaite et al., 2019, Gaafar et al., 2019, Lecoq et al., 2019).
Species demarcation criteria
The criteria demarcating species in the genus are:
• Less than 95% identity in the amino acid sequence of the RdRP
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| maize yellow stripe virus | RNA1: AJ969412*; RNA2: AJ696413*; RNA3: AJ969414*; RNA4: AJ969415; RNA5: AJ969416 | MYSV |
* Sequences do not comprise the complete genome segment.
Virus names and virus abbreviations are not official ICTV designations.

