Family: Phenuiviridae
Genus: Tenuivirus
Distinguishing features
Tenuiviruses infect a wide range of monocotyledonous plants and are transmitted from plant to plant by planthoppers or leafhoppers in which they also replicate and are transmitted transovarially. The eight viruses assigned to the genus Tenuivirus have genomes of 4–6 segments that encode two structural proteins [large protein (L) and nucleocapsid protein (N)], and 5–10 non-structural proteins including the viral movement protein (MP) that enables cell-to-cell movement in plant hosts, and the viral suppressor of RNA silencing (VSR) that counteracts plant antiviral defence mechanisms. Tenuiviruses have a relatively complex genome structure compared to other members of the family Phenuiviridae, with four to six genome components. The apparent lack of a membrane-bound virus particle distinguishes tenuiviruses from some other viruses in the family Phenuiviridae. Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences, viruses classified in the genus Tenuivirus form a monophyletic cluster clearly distinguished from other phenuivirids (Ramírez and Haenni 1994, Kormelink et al., 2021, Ramirez and Haenni 2021).
Virion
Morphology
Virions have a thin filamentous shape 2–2.5 nm in diameter with lengths (200–3,000 nm) proportional to the sizes of the segmented genomic RNAs. The filamentous particles may appear to be spiral-shaped, branched or panhandled (Figure 1 Phenuiviridae). No envelope has been observed (Koganezawa 1977, Ishikawa et al., 1989).
Physicochemical and physical properties
Purified preparations of rice stripe virus (RStV) can be separated into four or five components by sucrose density gradient centrifugation but form one component with a buoyant density 1.282–1.288 g cm−3 when centrifuged to equilibrium in CsCl solutions (Gingery et al., 1981, Hibino et al., 1985b, Ishikawa et al., 1989). Not only viral negative-sense genome RNAs (vRNAs) but also a substantial amount of positive-sense, virus genome complementary RNAs, known as antigenome RNAs (vcRNA), are encapsidated and form virions. Small amounts of a minor 330 kDa protein are co-purified. This protein has RNA polymerase activity and is capable of replicating and transcribing the RNA segments in vitro (Toriyama 1986, Toriyama 1987, Toriyama and Watanabe 1989, Ramírez and Haenni 1994).
Nucleic acid and Protein
The tenuivirus genome encompasses four to six single-stranded segments of negative-sense or ambisense RNA. The terminal nucleotides of each segment occur in a canonical, conserved sequence (in coding sense) 5′-ACACAAAGUC… GACUUUGUGU-3′ and may form panhandle structures typical of other members of the class Bunyaviricetes (Table 2 Phenuiviridae). RNA1 (9.1–9.8 kb) encodes a protein with a predicted molecular mass of 336–341 kDa that is homologous with the bunyaviral RNA-directed RNA polymerase (RdRP) domain. RNA2 (1.8–4.1 kb) encodes two non-structural proteins of 91–109 kDa and 32–36 kDa. RNA3 (1.6–3.1 kb) and the functionally homologous RNA5 (2.7 kb) of rice grassy stunt virus (RGSV), encodes a protein of 33–36 kDa that is homologous with the tenuivirus/phlebovirus N domain, as well as a non-structural protein of 22–24 kDa. RNA4 (1.6–2.2 kb) and the functionally homologous RGSV RNA6 (2.6 kb) encode non-structural proteins of 32–36 kDa that share sequence homology and/or structural characteristics with the MP of plant viruses, as well as a non-structural protein of 22–24 kDa (Table 3 Phenuiviridae) (Toriyama et al., 1998, Sõmera et al., 2020, Kormelink et al., 2021, Ramirez and Haenni 2021).
Genome organization and replication
The tenuivirus genome encompasses four to six single-stranded segments of negative-sense or ambisense RNAs (Figure 1 Tenuivirus). The 3′- and 5′-terminal sequences of each single-stranded RNA are largely complementary over about 20 bases. The genomic RNA 5′- and 3′-ends can base-pair, and probably give rise to RNPs. Genomic RNAs are not modified at the 5′- and 3′-ends. Viral mRNAs are not polyadenylated and are truncated relative to the vRNA; a 5′-methylated cap is derived from host mRNA via ‘cap snatching’ mediated by the endonuclease function of the L protein. Several RNA segments encode two proteins in an ambisense arrangement. These ORFs are separated by a noncoding intergenic region that potentially forms a long A/U rich stem-loop structure. RNA1 of all tenuiviruses encodes one large protein (L protein) including an RdRP domain on vcRNA1. RNA1 of RGSV and European wheat striate mosaic virus (EWSMV) encode an additional non-structural protein of unknown function in ambisense orientation (Takahashi et al., 1990, Shimizu et al., 1996, Toriyama et al., 1998, Sõmera et al., 2020). For all tenuiviruses, RNA2 encodes two non-structural proteins (NSvc2 on vcRNA2 and NSv2 on vRNA2) in ambisense orientation that appear to be homologous among tenuiviruses. The RStV NSvc2 is cleaved into two proteins, like Gn and Gc of phenuivirids; the uncleaved proteins mature from ER to the Golgi complex via COPII-dependent vesicle transport and act as helper components in insect transmission. These proteins also show some similarity to the glycoproteins encoded by the M segment of uukuviruses (Yao et al., 2014, Lu et al., 2019, Kormelink et al., 2021, Xu et al., 2021). The NSv2 of RStV and RGSV are viral suppression of RNA silencing (VSR) that show weak activities in the suppression of RNA silencing (Du et al., 2011, Nguyen et al., 2015). RNA3 and the functionally homologous RNA5 of RGSV (Toriyama et al., 1998) encodes two proteins: N on vcRNA3 and VSR on vRNA3 (Hemmes et al., 2007, Xiong et al., 2009, Netsu et al., 2015). RNA4 and the functionally homologous RNA6 of RGSV encode two non-structural proteins, MP on vcRNA4 that plays an important role in the viral cell-to-cell movement of plant hosts, and a major non-capsid protein (NCP) on vRNA4 that forms characteristic inclusion bodies in infected plant and/or vector insect cells and is presumably associated with symptom development of infected plants (Gingery et al., 1981, Toriyama et al., 1998, Xiong et al., 2008, Hiraguri et al., 2011, Xu and Zhou 2012, Xu et al., 2021).
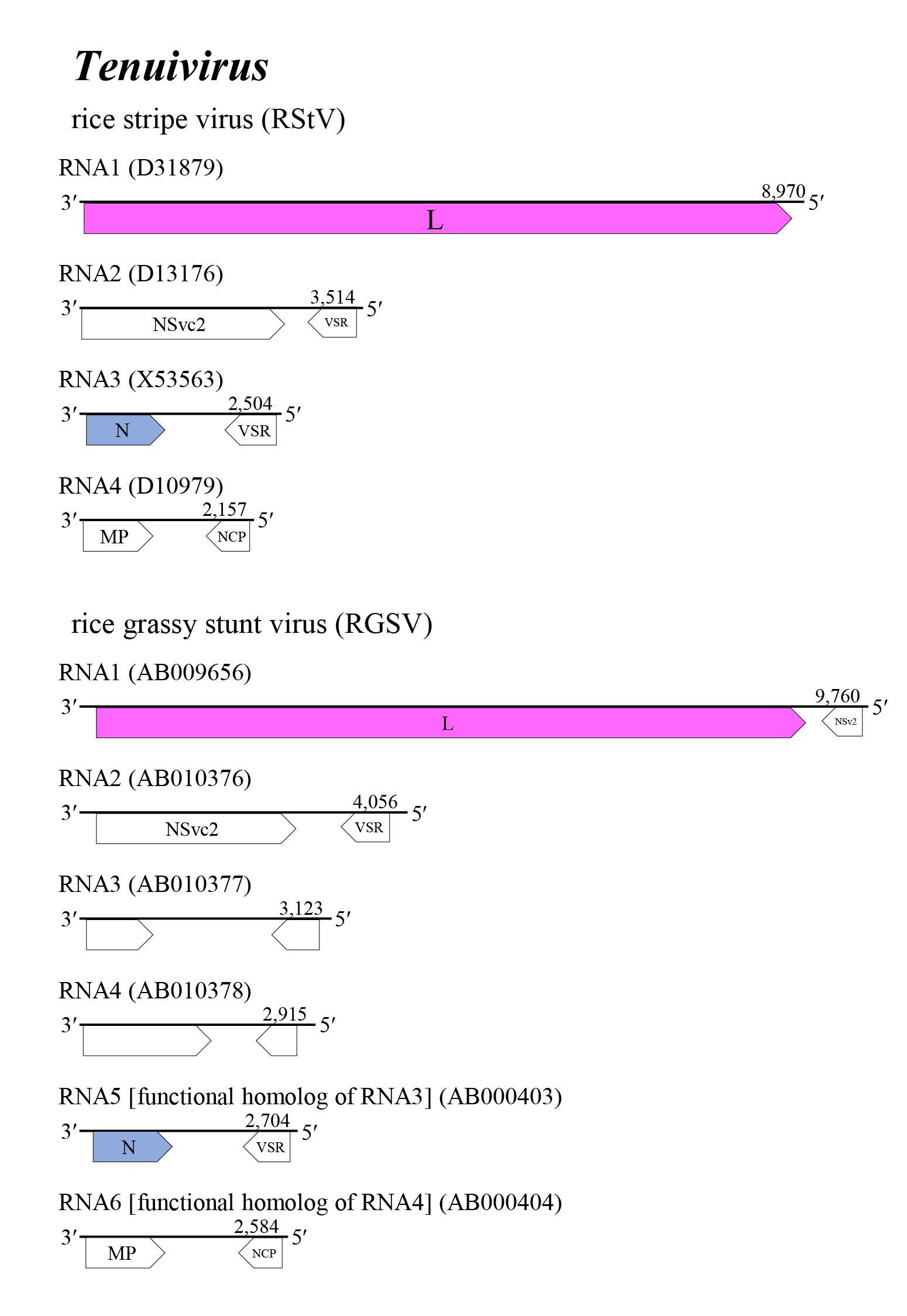 |
| Figure 1 Tenuivirus. Genome organization of some tenuiviruses. Coloured boxes depict ORFs that encode N, nucleocapsid protein and L, large protein. White boxes depict ORFs that encode MP, non-structual cell-to-cell movement protein; NCP, major non-capsid protein; VSR, viral suppressor of RNA silencing; and NSvc2, non-structural proteins translated from the complementary to the virion RNA2. |
Biology
Host range
Tenuiviruses infect monocotyledonous plants of the family Poaceae: barley [Hordeum vulgare (L., 1753)], maize [Zea mays (L., 1753)], millet [Eleusine coracana ((L.) Gaertn, 1788), Panicum miliaceum (L., 1753), Pennisetum glaucum ((Maire) Brunken, 1977), and Setaria italica ((L.) P.Beauv., 1812)], oat [Avena sativa (L., 1753)], rice [Oryza sativa (L., 1753)], rye [Secale cereale (L., 1753)], and wheat [Triticum aestivum (L., 1753)]. Outbreaks of tenuivirus diseases can become serious problems of rice and maize production. Stripe disease caused by RStV was first described in Japan in 1897. RStV occurs frequently in major rice-producing regions of many Eastern Asian countries such as China, Korea and Japan. Grassy stunt diseases caused by RGSV was first discovered in the Philippines in 1963 and occurs in major rice-producing regions of Southern, South-eastern and Eastern Asia. Hoja blanca disease caused by rice hoja blanca virus (RHBV) has occurred in Colombia since the 1930s and broke out in Cuba and Venezuela in the mid-1950s. Stripe disease caused by maize stripe virus (MStV) was first reported in the Mauritius Islands in 1929 and by 1992 was affecting 80% of crops. MStV occurs in major maize-producing regions of in Central and South America, Africa and Australia (Gingery et al., 1981, Hibino et al., 1985a, Hibino 1996, Martin et al., 2020, Xu et al., 2021).
Transmission
Tenuiviruses are transmitted by planthoppers or leafhoppers of specific species in a circulative and propagative manner. The major vectors are insects of species Caenodelphax teapae (Fowler, 1905) [Urochloa hoja blanca virus (UHBV)], Javesella pellucida (Fabricius) [EWSMV], Laodelphax striatellus Fallén [RStV], Nilaparvata lugens Muir [RGSV], Peregrinus maidis Ashmead [MStV], Tagosodes orizicolus Muir [RHBV], T. cubanus Muir [Echinochloa hoja blanca virus (EHBV)], and Ukanodes tanasijevici Dlabola [Iranian wheat stripe virus (IWSV)]. Tenuiviruses can be transmitted transovarially by virus-infected female planthoppers to their offspring, and through sperm from virus-infected males. Mechanical transmission using sap extracts is possible with RStV (Gingery et al., 1981, Falk and Tsai 1998, Xu and Zhou 2012, Sõmera et al., 2020).
Cytopathic effects
Characteristic inclusion bodies, consisting almost entirely of NCP encoded on vRNA4 of RStV and MStV, are formed in cells of infected plants. Similarly, NCP encoded on vRNA6 of RGSV accumulates in large amounts in both infected plants and vector insects (Gingery et al., 1981).
Antigenicity
The N proteins of MStV, RStV, RGSV and RHBV are serologically related; the NCP of RStV and RGSV are related. Likewise, the N proteins of RHBV, EHBV and UHBV are serologically related (Hibino et al., 1985b, Falk et al., 1987).
Species demarcation criteria
The criteria demarcating species in the genus are:
• Less than 95% identity in the amino acid sequence of the RdRP
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| black spruce virus | RNA3: AF177865* | BSV |
| wheat yellow head virus# | WYHV |
* Sequences do not comprise the complete genome segment.
Virus names and virus abbreviations are not official ICTV designations.

