Family: Phenuiviridae
Genus: Goukovirus
Distinguishing features
Five viruses, Aphalara polygoni bunya-like virus (ApBLV), Ceraphron bunya-like virus (CerBLV), Cumuto virus (CUMV), Gouléako virus (GOLV) and Yíchāng insect virus (YIV) are assigned to the genus Goukovirus. The goukovirus genome has four genes, encoding a large protein (L), two external glycoproteins (Gn and Gc), and a nucleocapsid protein (N). Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences, viruses classified in the genus Goukovirus form a monophyletic cluster clearly distinguished from other phenuivirids. Goukovirus RNA has been found in pools of the aphid Aulacorthum magnoliae (Essig & Kuwana, 1918), of the jumping plant louse Aphalara polygoni (Förster, 1848) and of the mosquito Culex portesi (Senevet & Abonnenc, 1941) (Marklewitz et al., 2011, Auguste et al., 2014, Li et al., 2015).
Virion
Morphology
Virions are spherical or pleomorphic, 40–60 nm in diameter, and have surface glycoprotein projections of 5–10 nm which are embedded in a lipid bilayer envelope approximately 5 nm thick (Marklewitz et al., 2011, Auguste et al., 2014).
Nucleic acid and Protein
The goukovirus genome encompasses three segments of negative-sense RNA. The terminal nucleotides of each segment occur in a canonical, conserved sequence (in coding sense) 5′-ACACAGUGAC…GACUUUGUGU-3′ and may form panhandle structures similar to those of other members of the class Bunyaviricetes (Table 2 Phenuiviridae). The L segment (6.4 kb) encodes a protein with a predicted molecular mass of 238–240 kD that includes a region that shares homology with the bunyaviral RNA-directed RNA polymerase (RdRP) domain. The M segment (3.2–3.3 kb) encodes a protein of 107–110 kDa that shares homology with phlebovirus glycoprotein G1 and G2 sequences, and also to the phlebovirus glycoprotein C-terminal Ig-like domain. The S segment (1.0–1.2 kb) encodes a protein of 28–29 kDa that shares homology with the tenuivirus/phlebovirus N domain (Table 3 Phenuiviridae).
Genome organization and replication
Goukovirus genome arrangement is similar to that of citriciviruses, beidiviruses, hudiviruses, hudoviruses, phasiviruses, pidchoviruses and tanzaviruses (Figure 1 Goukovirus). All three genomic RNAs contain untranslated regions flanking a single ORF which, based on comparisons with other negative-sense RNA viruses, is predicted to be contained in the virus complementary strand. The L, M, and S segments putatively encode L, a glycoprotein precursor (comprising Gn and Gc) and N, respectively. The Gn and Gc glycoproteins of 53–56 kDa and 52–54 kDa, respectively, were earlier referred to as G1 and G2 based on their apparent size following gel electrophoresis. The goukovirus genome does not encode either of the non-structural proteins NSm or NSs. Details of virus replication are unknown (Marklewitz et al., 2011, Auguste et al., 2014).
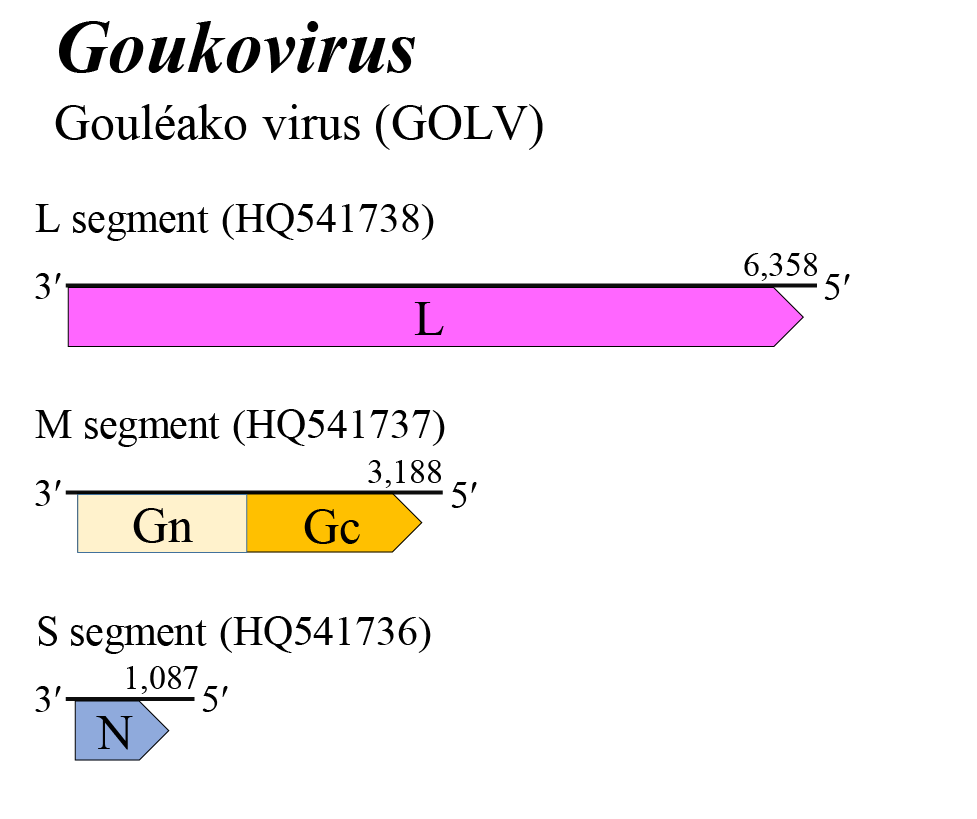 |
| Figure 1 Goukovirus. Genome organization of a goukovirus. Coloured boxes depict ORFs that encode N, nucleocapsid protein; Gn and Gc, external glycoproteins; and L, large protein. |
Biology
Goukoviruses have been found in aphids, jumping plant lice, and mosquitoes. The viruses replicate in insects but not in vertebrate cells (Marklewitz et al., 2011, Auguste et al., 2014, Li et al., 2015).
Species demarcation criteria
The criteria demarcating species in the genus are:
• Less than 95% identity in RdRP amino acid sequence.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Diaporthe gulyae goukovirus 1 | L: OR224974 | DiGGV1 |
| Sefomo virus | L: MZ202295; M: MZ202296; S: MZ202297 | SeV |
| Trichopria drosophilae bunya-like virus | L: BK061543 | TDBLV |
Virus names and virus abbreviations are not official ICTV designations.

