Family: Phenuiviridae
Genus: Rubodvirus
Distinguishing features
Rubodviruses were discovered first in apple and later in grapevine. Four viruses are assigned to the genus Rubodvirus. The rubodvirus genome has three genes, encoding a large protein (L), a nucleocapsid protein (N) and a putative viral movement protein (MP) that may enable cell-to-cell movement in plant hosts. Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences, viruses classified in the genus Rubodvirus form a monophyletic cluster clearly distinguished from other phenuivirids (Rott et al., 2018, Diaz-Lara et al., 2019).
Virion
Virion morphology is unknown. Based on the putative proteins encoded by the virus genome, the virion is probably a filamentous virion without an envelope.
Nucleic acid and Proteins
The rubodvirus genome encompasses three segments of negative-sense RNA: RNA1 (7.2–8.1 kb), RNA2 (1.5–1.6 kb), and RNA3 (1.3–1.6 kb). The terminal nucleotides of each segment occur in a canonical, conserved sequence (in coding sense) 5′-ACCCCUCCAA…UGGAGGGGGU-3′ and may form panhandle structures typical of other members of the class Bunyaviricetes (Table 2 Phenuiviridae). All three genomic RNAs contain untranslated regions flanking a single ORF which, based on comparisons with other negative-sense RNA viruses, is predicted to be contained in the viral complementary strand. In silico analysis of rubodvirus putative ORF sequences suggests that rubodviruses encode two structural proteins: L with a predicted molecular mass of 276–304 kDa that is homologous with the bunyaviral RNA-directed RNA polymerase (RdRP) domain, and N of 32 kDa that is homologous with the tenuivirus/phlebovirus N domain, as well as one non-structural protein of 44–45 kDa that is homologous with and/or shares structural characteristics with the MP of other viruses (Table 3 Phenuiviridae) (Rott et al., 2018, Diaz-Lara et al., 2019).
Genome organization and replication
The rubodvirus genome consists of three negative-sense RNA segments (Figure 1 Rubodvirus). RNA1, RNA2, and RNA3 putatively encode an RNA-directed RNA polymerase (RdRP), an MP (instead of external glycoproteins) and N, respectively. Most plant viruses encode viral suppressor proteins that counteract plant antiviral defense mechanisms through RNA silencing. There is a possibility that one or more of the three proteins encoded by rubodviruses may interfere with RNA silencing. Multifunctional roles have been reported for viral suppressor proteins of other viruses (Rott et al., 2018, Diaz-Lara et al., 2019, Kormelink et al., 2021).
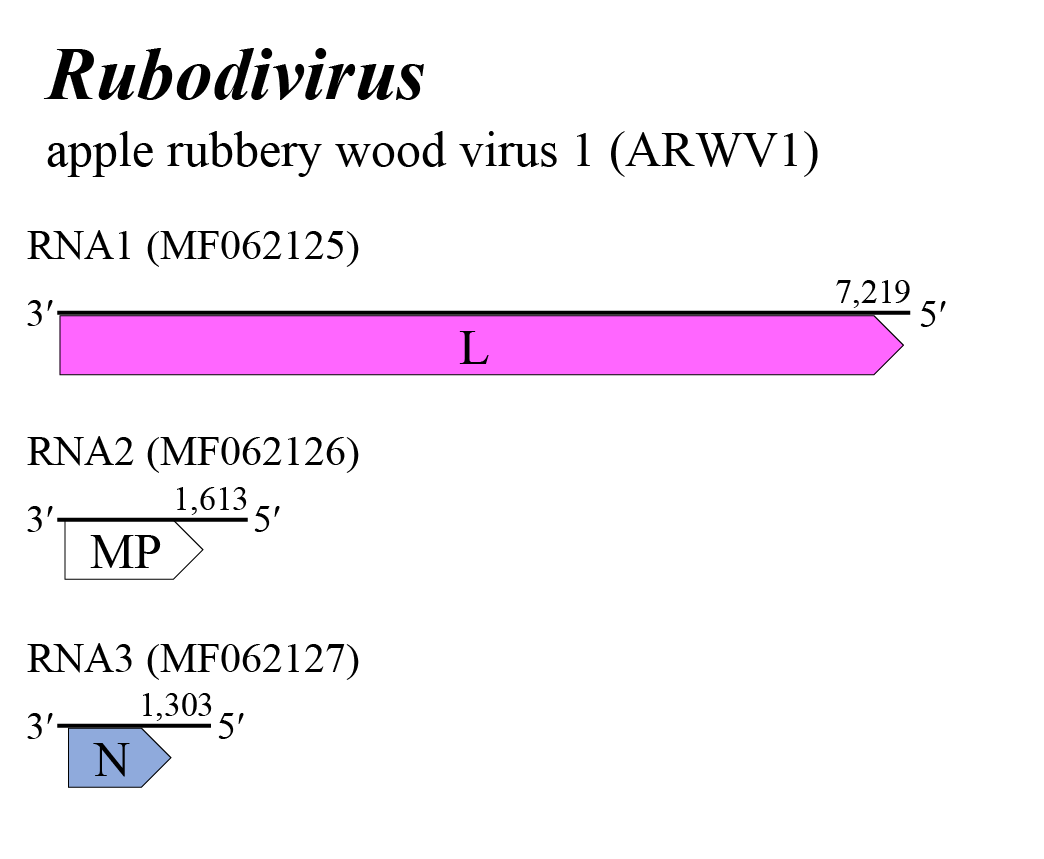 |
| Figure 1 Rubodvirus. Genome organization of a rubodvirus. Coloured boxes depict ORFs that encode N, nucleocapsid protein and L, large protein. A white box depicts an ORF that encodes MP, non-structural cell-to-cell movement protein. |
Biology
Hosts of rubodviruses are plants of the families Vitaceae: grapevine [Vitis vinifera (L.,1758)] and Rosaceae: apple [Malus domestica (Suckow) Borkh., 1803)], pear [Pyrus pyrifolia ((Burm.f.) Nakai, 1926)], and quince [Cydonia oblonga (Mill, 1768)]. Viruses are transmitted by grafting (McCrum et al., 1960, Rott et al., 2018, Diaz-Lara et al., 2019).
Species demarcation criteria
The criteria demarcating species in the genus is:
• Less than 95% identity in the amino acid sequence of the RdRP
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Qīngdǎo RNA virus 3 | RNA1: LC726787; RNA2: LC726788 | QRV3 |
Virus names and virus abbreviations are not official ICTV designations..

