Family: Phenuiviridae
Genus: Horwuvirus
Distinguishing features
Three viruses, Fitzroy Crossing tenui-like virus 1 (FCTenV1), Solenopsis invicta virus 14 (SINV14), and Wǔhàn horsefly virus (WhHV) are assigned to the genus Horwuvirus. Horwuvirus RNA has been found by high-throughput sequencing of RNA from fire ants [Solenopsis invicta (Buren, 1972)] in USA, horseflies (Tabanidae spp.) in China, and mosquitoes [Culex annulirostris (Skuse, 1889)] in Australia. The horwuvirus genome has five genes, encoding a large protein (L), two external glycoproteins (Gn and Gc), a nucleocapsid protein (N) and a non-structural protein (NSs), a similar arrangement to that of wenriviruses. Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences, viruses classified in the genus Horwuvirus form a monophyletic cluster clearly distinguished from other phenuivirids. There currently is no cultured horwuvirus isolate (Li et al., 2015, Williams et al., 2020).
Virion
Morphology
Virion morphology is unknown. Based on the putative proteins encoded by the virus genome, the virion is probably a spherical or pleomorphic virion with the enveloped structure.
Nucleic acid and Protein
The horwuvirus genome encompasses four segments of negative-sense RNA, namely L (8.5–9.5 kb), M (2.2–2.8 kb), S (0.9–1.9 kb) and S2 (0.9–1.7 kb). All four genomic RNAs contain untranslated regions flanking a single ORF which, based on comparisons with other negative-sense RNA viruses, is predicted to be contained in the virus-complementary strand. Analysis of the terminal sequences has yet to be completed. In silico analysis of horwuvirus putative ORF sequences suggests that horwuviruses encode four structural proteins: L with a predicted molecular mass of 332–363 kDa, a glycoprotein precursor (GP) of 84–94 kDa and N of 35–52 kDa, all of which share sequence homology and/or structural characteristics with the cognate proteins of other phenuivirids (Table 3 Phenuiviridae) (Li et al., 2015, Williams et al., 2020).
Genome organization and replication
The horwuvirus genome consists of four negative-sense RNAs (Figure 1 Horwuvirus). The L, M, and S segments putatively encode L, a glycoprotein precursor (comprising Gn and Gc) and N, respectively. The Gn and Gc proteins of 33–44 kDa and 50–52 kDa were earlier referred to as G1 and G2 based on apparent size following gel electrophoresis. In addition, the horwuvirus S2 segment, like that of wenriviruses, encodes a non-structural protein (NSs2) of 33–41 kDa of unknown function. Details of virus replication are unknown (Li et al., 2015, Williams et al., 2020).
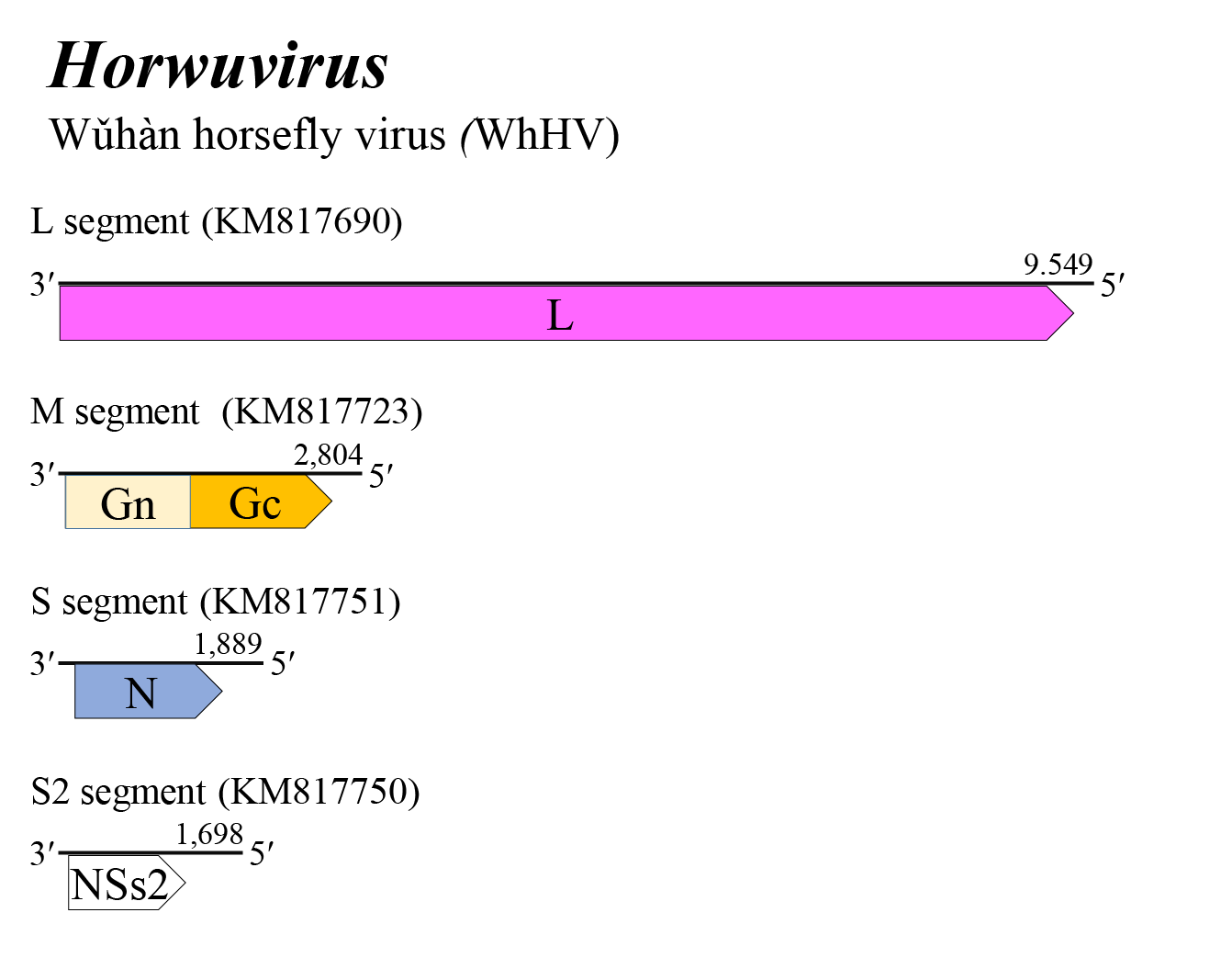 |
| Figure 1 Horwuvirus. Genome organization of a horwuvirus. Coloured boxes depict ORFs that encode N, nucleocapsid protein; Gn and Gc, external glycoproteins; and L, large protein. A white box depicts an ORF that encodes NSs2, non-structural protein. |
Biology
Horwuviruses have been detected in fire ants, horseflies and mosquitoes (Li et al., 2015, Williams et al., 2020).
Species demarcation criteria
The criteria demarcating species in the genus are:
• Less than 95% identity in RdRP amino acid sequence.

