Family: Phenuiviridae
Genus: Entovirus
Distinguishing features
Entoleuca phenui-like virus 1 (EnPLV1) is assigned to Entovirus entoleucae, the only species in the genus. Entovirus RNA has been detected by high-throughput sequencing of RNA from a fungus of the genus Entoleuca obtained from the rhizosphere of avocado in Spain. The entovirus genome has three genes, encoding two structural proteins, a large protein (L) and a nucleocapsid protein (N), and one non-structural gene likely encoding a viral movement protein (MP) that enables cell-to-cell movement in fungal hosts. Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences, viruses classified in the genus Entovirus form a monophyletic cluster clearly distinguished from other phenuivirids (Velasco et al., 2019).
Virion
Morphology
Virion morphology is unknown. Based on the putative proteins encoded by the virus genome, the virion is probably a filamentous virion with the non-enveloped structure.
Nucleic acid and Protein
The entovirus genome consists of two single-stranded RNA segments: RNA1 (7.3 kb) and RNA2 (2.8 kb). The terminal nucleotides of each segment occur in a canonical, conserved sequence (in coding sense) 5′-ACACAAAGAC… GUCUUUGUGU-3′ that may form panhandle structures typical of other members of the class Bunyaviricetes (Table 2 Phenuiviridae). In silico analysis of entovirus putative ORF sequences suggests that the entovirus genome encodes two proteins with predicted molecular masses of 271 kDa and 39 kDa that share homology with the bunyaviral RNA-directed RNA polymerase (RdRP) domain and the tenuivirus/phlebovirus N domain, respectively, and one predicted protein of 45 kDa that shares sequence homology and/or structural characteristics with the MP of plant viruses (Table 3.Phenuiviridae) (Velasco et al., 2019).
Genome organization and replication
Entovirus genome arrangement is similar to that of coguviuses and lentinuviruses (Figure 1 Entovirus). The entovirus genome encompasses two segments of negative-sense RNA: RNA1, encoding the L protein that is the putative RdRP, and the ambisense RNA2, which is transcribed by RdRP to form a subgenomic virus-complementary mRNA that encodes N from a viral antigenome RNA2, and also transcribed to form a subgenomic virus-sense mRNA that encodes MP from a viral genome RNA2 (Figure 1 Entovirus). The two ORFs on RNA2 are separated by a noncoding intergenic region that potentially forms a long A/U rich stem-loop structure. Details of virus replication are unknown (Velasco et al., 2019, Rodriguez Coy et al., 2022).
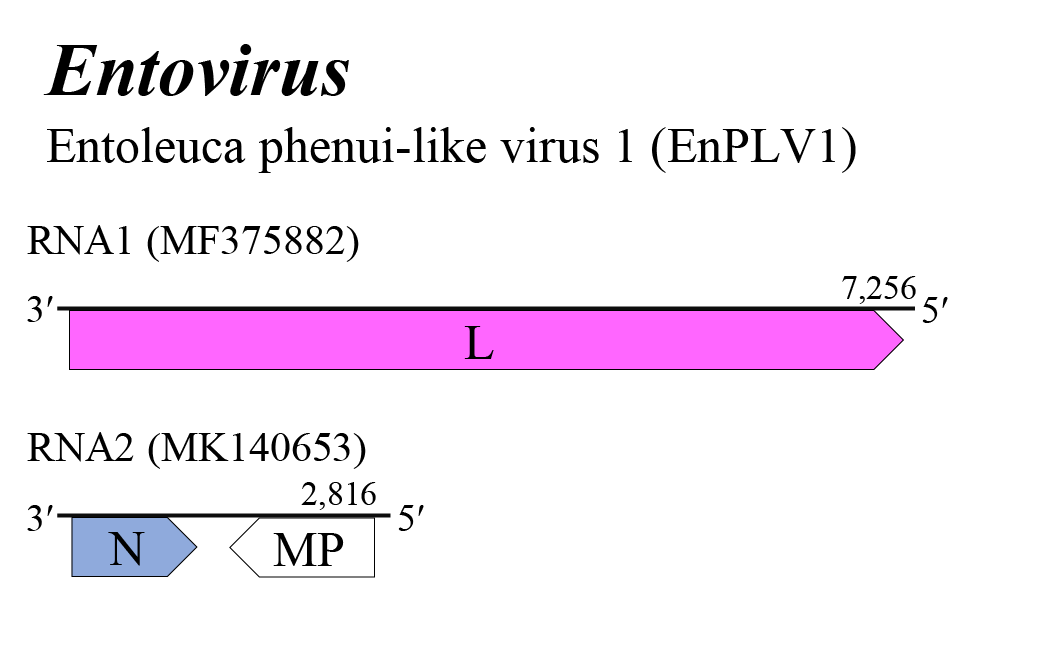 |
| Figure 1 Entovirus. Genome organization of an entovirus. Coloured boxes depict ORFs that encode N, nucleocapsid protein and L, large protein. A white box depicts an ORF that encodes MP, non-structural cell-to-cell movement protein. |
Biology
The only known isolate was detected in a fungus of the genus Entoleuca obtained from the rhizosphere of avocado (Velasco et al., 2019).
Species demarcation criteria
Not defined as the genus currently includes only a single species.

