Family: Phenuiviridae
Takahide Sasaya (笹谷孝英), Gustavo Palacios, Thomas Briese, Francesco Di Serio, Martin H. Groschup, Yutaro Neriya (煉谷裕太朗), Jin-Won Song (송진원), and Yasuhiro Tomitaka (冨髙保弘)
The citation for this ICTV Report chapter is the summary published as :
Corresponding authors: Takahide Sasaya (笹谷孝英) ([email protected]); Gustavo Palacios ([email protected])
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: September 2023, updated June 2024
Summary
The family Phenuiviridae includes 23 genera and 159 species for 180 viruses with segmented negative-sense or ambisense RNA genomes of 8.1–25.1 kb in total (Table 1 Phenuiviridae). Virions are typically enveloped with spherical or pleomorphic morphology, but non-enveloped filamentous virions have also been reported. Individual RNA genome segments have partially complementary termini. Phenuivirid genomes generally have four genes encoding structural proteins (L, Gn, Gc, and N). However, several phenuivirid genomes lack genes for Gn and Gc and many phenuivirid genomes encode additional non-structural proteins, such as as an interferon response inhibitor protein that impacts interferon production in host cells and viral virulence, an apoptosis inhibitor protein that targets to the mitochondrial outer membrane and exerts an antiapoptotic function, a movement protein (MP) that enables cell-to-cell movement in plant and fungus hosts and a viral suppressor protein of RNA silencing (VSR) that counteracts host antiviral defence mechanisms, in additional genes or in alternative open reading frames. The family is ecologically diverse with members infecting humans and livestock animals, birds, crustaceans, plants and fungi. Phenuivirids have also been detected in invertebrates, including arthropods, some of which act as biological vectors for transmission to other animals or plants, or may serve as their unique host. Phenuivirids include important pathogens of humans and livestock, and have an impact on seafoods and agricultural crops.
Table 1 Phenuiviridae. Characteristics of members of the family Phenuiviridae
| Characteristic | Description |
| Example | Rift Valley fever virus (RVFV) [S segment: DQ380151; M segment: DQ380206; L segment DQ375403], species Phlebovirus riftense, genus Phlebovirus |
| Virion | Enveloped spherical or pleomorphic virions, 80–120 nm in diameter comprised of a helical nucleocapsid surrounded by a lipid bilayer envelope. Some phenuivirids produce non-enveloped filamentous virions, 2–2.5 nm in diameter, 200–3000 nm in length |
| Genome | 8.1–25.1 kb in total, comprising 2–8 segments of negative-sense or ambisense RNA, of 0.8–9.8 kb |
| Replication | Cytoplasmic. The nucleocapsid protein (N) encapsidates the genomic RNA forming ribonucleoprotein (RNP) complexes with large protein (L) containing the viral RNA-directed RNA polymerase (RdRP). Anti-genomic RNAs are generated and serve as templates for synthesis of nascent RNP complexes containing genomic RNA. |
| Translation | From capped mRNAs that lack poly(A) termini. The 5′-cap structure is derived from cellular mRNAs via cap-snatching |
| Host range | Vertebrates, including mammals and birds, invertebrates, plants, and fungi |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Bunyaviricetes, order Hareavirales: 23 genera and 159 species |
Genus Bandavirus. Viruses assigned to this genus infect mammals, including humans, and are transmitted by ticks in which they replicate and propagate by transovarian transmission. Several bandaviruses cause fever and signs of central nervous system involvement in young ruminants and/or have been associated with fatal thrombocytopenia fever syndrome in humans. Virus particles are spherical or pleomorphic and have surface glycoprotein projections which are embedded in a lipid bilayer envelope. Genomes consist of three segments containing four canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N, and one non-structural protein gene (NSs).
Genus Beidivirus. The single virus assigned to this genus was found by high-throughput sequencing of RNA from a pool of flies from China. The genome consists of three segments and contains the canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N.
Genus Bocivirus. Viruses assigned to this genus have been found in RNA from plant-pathogenic fungi. Genomes consist of three or two segments containing two structural protein genes encoding L and N, and one non-structural protein gene likely encoding a cell-to-cell movement protein (MP) that enables cell-to-cell movement in plant and fungus hosts.
Genus Citricivirus. The single virus assigned to this genus was found by high-throughput sequencing of RNA from a brown citrus aphid pool from China. The genome consists of three segments and contains the canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N.
Genus Coguvirus. Viruses assigned to this genus infect a wide range of dicotyledon plants and are transmitted by grafting. Virus particles are flexuous filaments and do not have envelopes. Genomes consist of two segments with two structural protein genes encoding L and N, and one non-structural protein gene probably associated with MP.
Genus Entovirus. The single virus assigned to this genus was found by high-throughput sequencing of RNA from entoleucan fungi obtained from the rhizosphere of avocados in Spain. The genome consists of two segments with two structural protein genes encoding L and N, and one non-structural protein gene likely encoding MP.
Genus Goukovirus. Viruses assigned to this genus have been isolated from, or detected in, mosquitoes and aphids. Goukoviruses replicate in insects but not in vertebrate cells. Virus particles are spherical or pleomorphic and have surface glycoprotein projections which are embedded in a lipid bilayer envelope. Genomes consist of three segments containing four canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N.
Genus Horwuvirus. Viruses assigned to this genus have been detected by high-throughput sequencing of RNA from fire ants, horseflies, and mosquitoes. Genomes consist of four segments and contain four canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N, and one non-structural protein gene of unknown function.
Genus Hudivirus. The single virus assigned to this genus was found by high-throughput sequencing of RNA from a fly pool from China. The genome consists of three segments and contains four canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N.
Genus Hudovirus. The single virus assigned to this genus was found by high-throughput sequencing of RNA from a butterfly pool from China. The genome consists of three segments and contains four canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N.
Genus Ixovirus. Viruses assigned to this genus have been detected in RNA from ticks. Genomes consist of two segments and contain only two structural protein genes encoding L and N. Vertebrate hosts have not been identified.
Genus Laulavirus. Viruses assigned to this genus have been found in RNA from fungi and ticks. Genomes consist of three segments containing two structural protein genes encoding L and N, and one non-structural protein gene probably associated with the MP of plant viruses.
Genus Lentinuvirus. The single virus assigned to this genus was found by high-throughput sequencing of RNA from shiitake from Japan. The genome consists of two segments containing two structural protein genes encoding L and N, and one non-structural protein gene likely encoding MP.
Genus Mechlorovirus. Viruses assigned to this genus infect a wide range of dicotyledonous and monocotyledonous plants are are transmitted by planthoppers. Virus particles are filamentous and flexuous and do not have envelopes. This virus has the most complex genome organization known among phenuivirids, with eight segments containing thirteen genes encoding L, N, and eleven non-structural proteins of unknown function.
Genus Mobuvirus. Viruses assigned to this genus have been found by high-throughput sequencing of RNA from fleas, moths and mosquitoes. Genomes consist of three segments and contain the canonical phenuivirid structural protein genes encoding L, glycoprotein precursor, and N. Mobuviruses of moths encode an additional non-structural protein of unknown function.
Genus Phasivirus. Viruses assigned to this genus have been isolated from flies and mosquitoes. Phasiviruses replicate in insects but not in vertebrate cells. Virus particles are spherical or pleomorphic and have surface glycoprotein projections which are embedded in a lipid bilayer envelope. Genomes consist of three segments containing four canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N.
Genus Phlebovirus. Viruses assigned to this genus infect mammals, including humans, and are transmitted by mosquitoes, phlebotomine sandflies and ticks in which they replicate and propagate by transovarian transmission. Several phleboviruses cause abortion and often manifest as congenital malformations in livestock and/or have been associated with fatal febrile illness and occasionally encephalitides in humans. Virus particles are spherical or pleomorphic and have surface glycoprotein projections which are embedded in lipid bilayer envelopes. Genomes consist of three segments containing four canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N, and two non-structural protein genes encoding NSm and NSs.
Genus Pidchovirus. Viruses assigned to this genus have been found by high-throughput sequencing of RNA from beetles and moths. Genomes consist of three segments containing four canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N.
Genus Rubodvirus. Viruses assigned to this genus have been found in apples and grapevines and are transmitted by grafting. Genomes consist of three segments containing two structural protein genes encoding L and N, and one non-structural protein gene likely encoding MP.
Genus Tanzavirus. The single virus assigned to date to this genus has been detected by high-throughput sequencing of a plasma sample from a human in Tanzania with a history of fever, headache and back pain. The genome consists of three segments containing four canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N.
Genus Tenuivirus. Viruses assigned to this genus infect a wide range of monocotyledon plants and are transmitted by planthoppers or leafhoppers in which they replicate and propagate by transovarian transmission. Virus particles are flexuous filaments and do not have envelopes. Several tenuiviruses cause serious problems in rice and maize production. Genomes have a relatively complex organization among phenuivirids with up to six segments and up to twelve genes encoding L, N, and five to ten non-structural proteins including MP, a viral suppressor of RNA silencing (VSR) that counteracts plant antiviral defence mechanisms, and a major non-capsid protein (NCP) that forms characteristic inclusion bodies in infected plant and/or vector insect cells.
Genus Uukuvirus. Viruses assigned to this genus infect birds and mammals, including humans, and are transmitted by ticks in which they replicate and propagate by transovarian transmission. Uukuviruses have not been considered to be of public health or agricultural significance, although antibodies to some uukuviruses have been detected in sera from humans, cattle, birds, and wild animals. Virus particles are spherical or pleomorphic and have surface glycoprotein projections that are embedded in lipid bilayer envelopes. Genomes consist of three segments containing four canonical phenuivirid structural protein genes encoding L, Gn, Gc, and N, and one non-structural protein gene encoding NSs.
Genus Wenrivirus. The single virus assigned to this genus infects prawns and shrimps. The viral infection increases the mortality of the prawns and shrimps. The genome consists of four segments containing four structural protein genes encoding L, Gn, Gc, and N, and one non-structural protein gene of unknown function.
Virion
Morphology
Phenuivirids that produce enveloped particles mainly infect vertebrates and invertebrates. By contrast, phenuivirids that produce nonenveloped particles mainly infect plants and fungi, as well as their arthropod vectors. Enveloped particles are spherical or pleomorphic, 80–120 nm in diameter, and have surface glycoprotein projections of 5–10 nm that are embedded in lipid bilayer envelopes approximately 5 nm thick (Figure 1 Phenuiviridae). Envelopes are usually derived from cellular Golgi membranes, or on occasion, from cell-surface membranes. Nonenveloped filamentous particles are spiral-shaped, branched or like a panhandle, 2–2.5 nm in diameter, with lengths proportional to the lengths (200–3000 nm) of the segmented genomic RNAs (Figure 1.Phenuiviridae).
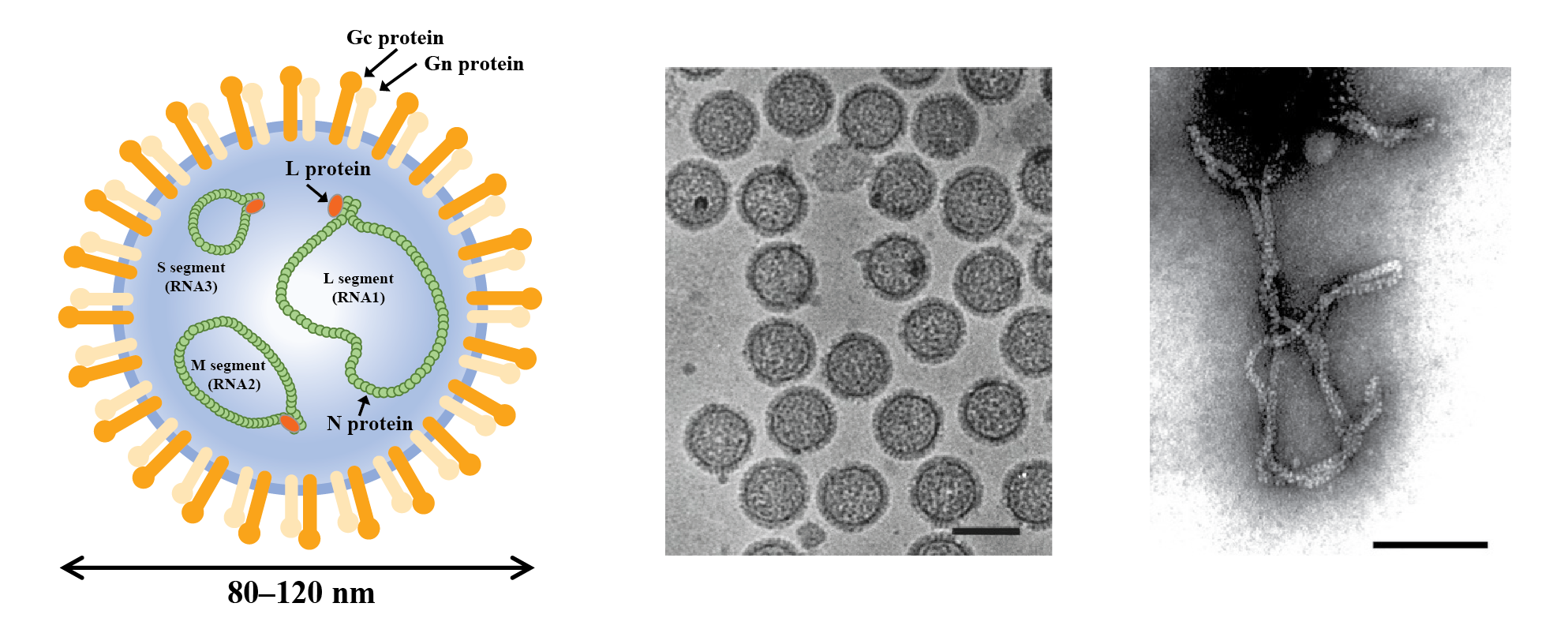 |
| Figure 1 Phenuivridae. (Left) Diagrammatic representation of an enveloped phenuivirid virion in cross-section. The surface spikes comprise two glycoproteins termed Gn and Gc (previously referred to as G1 and G2). The three helical nucleocapsids are panhandled in shape and comprise one each of the negative-sense RNA segments (L segment or RNA1, large or first; M segment or RNA2, medium or second; S segment or RNA3, small or third) encapsidated by N protein and associated with the L protein. (Middle) Cryo-electron micrograph of purified uukuvirus particles (Uukuniemi virus). The bar represents 100 nm (courtesy of C-H von Bornsdorff). (Right) Electron micrograph of purified tenuivirus particles (rice hoja blanca virus). The bar represents 100 nm (Courtesy of A.M. Espinoza). |
Physicochemical and physical properties
The relative molecular mass (Mr) of enveloped virions is 300×106 to 400×106 with an S20, W of 350–500. Virion buoyant densities in sucrose and CsCl are 1.16–1.18 and 1.20–1.21 g cm−3, respectively. Virions are sensitive to heat, lipid solvents, detergents, and formaldehyde. Non-enveloped virions are separated into four or five components by sucrose density gradient centrifugation, but form one component with a buoyant density 1.282–1.288 g cm−3 when centrifuged to equilibrium in CsCl solutions (Gingery et al., 1981, Hibino et al., 1985a, Ishikawa et al., 1989).
Nucleic acid
The viral genome comprises two to eight single-stranded molecules of negative-sense or ambisense RNA, which total 8.1–25.1 kb. The terminal nucleotides of each genome RNA segment are base-paired, forming non-covalently closed RNP complexes. The terminal sequences of genome segments are conserved among viruses of each genus but are different from those of viruses of other genera (Table 2 Phenuiviridae). The genomic RNAs are not modified at their 5′-termini. The Mr of the genome ranges from 4.8×106–8×106 and this constitutes 1–2% of the virion by weight. Viral mRNAs are not polyadenylated and are truncated relative to the genomic RNAs at their 3′-termini. The mRNAs have 5′-methylated caps and 10–18 non-templated nucleotides at their 5′-termini derived from host-cell mRNAs.
Table 2 Phenuiviridae. RNA segments of typical genus members
Genus | Virus | RNA segments | Conserved reverse complementary termini | |||||
|
| Number of segments | Number of bases |
|
| |||
|
|
| L or RNA1 | M or RNA2 | S or RNA3 (RNA2)* | S2 or RNA4 | 3′ | 5′ |
Bandavirus | severe fever with thrombocytopenia syndrome virus | 3 | 6368 | 3378 | 1744 | none | ACACAGAGAC…. | ….GUCUUUGUGU |
Beidivirus | Húběi diptera virus 3 | 3 | 6742 | 3763 | 1591 | none | ND | ND |
| Bocivirus | Botrytis cinerea bocivirus 1 | 3 | 6743 | 1653 | 1293 | none | ACACAAAGAU…. | ….AACUAUGUGU |
Citricivirus | Aphis citricidus bunyavirus | 3 | 7037 | 3462 | 1163 | none | ND | ND |
Coguvirus | citrus concave gum- associated virus | 2 | 6681 | none | 2703 | none | ACACAAAGAU…. | ….AUCUAUGUGU |
Entovirus | Entoleuca phenui-like virus 1 | 2 | 7256 | 2816 | none | none | ACACAAAGAC…. | ….GUCUUUGUGU |
Goukovirus | Gouléako virus | 3 | 6358 | 3188 | 1087 | none | ACACAGUGAC…. | ….GACUUUGUGU |
Horwuvirus | Wǔhàn horsefly virus | 4 | 9549 | 2804 | 1889 | 1698 | ND | ND |
Hudivirus | Húběi diptera virus 4 | 3 | 6947 | 4186 | 1236 | none | ND | ND |
Hudovirus | Húběi lepidoptera virus 1 | 3 | 7545 | 5114 | 1254 | none | ACACAAAGAC…. | ….GUCUUUGUGU |
Ixovirus | blacklegged tick virus 1 | 2 | 6733 | 2468 | none | none | ACACAAAGGC…. | ….GCCAUUGUGU |
Laulavirus | Laurel Lake virus | 3 | 7253 | 2506 | 1142 | none | ACACAAAGUC…. | ….GUCUUUGUGU |
Lentinuvirus | Lentinula edodes negative-strand RNA virus 2 | 2 | 7082 | none | 2754 | none | ACACAAAGAC…. | ….GUCUUUGUGU |
Mechlorovirus | melon chlorotic spot virus | 8 | 9096 | 1847 | 1598 | 1,592 | ACACAAAGUC…. | ….GACUUUGUGU |
Mobuvirus | Mothra virus | 3 | 6976 | 1967 | 1520 | none | ND | ND |
Phasivirus | Badu virus | 3 | 6863 | 3871 | 1513 | none | ACACAAAGAC…. | ….GUCUUUGUGU |
Phlebovirus | Rift Valley fever virus | 3 | 6404 | 3885 | 1690 | none | ACACAAAGAC…. | ….GUCUUUGUGU |
Pidchovirus | Pidgey virus | 3 | 6914 | 3850 | 1010 | none | ND | ND |
Rubodvirus | apple rubbery wood virus 1 | 3 | 7219 | 1613 | 1303 | none | ACCCCUCCAA…. | ….UGGAGGGGGU |
Tanzavirus | Dar es Salaam virus | 3 | 6474 | 3776 | 1309 | none | ND | ND |
Tenuivirus | rice stripe virus | 4 | 8970 | 3514 | 2504 | 2,157 | ACACAAAGUC…. | ….GACUUUGUGU |
Uukuvirus | Uukuniemi virus | 3 | 6423 | 3229 | 1720 | none | ACACAAAGAC…. | ….GUCUUUGUGU |
Wenrivirus | Mourilyan virus | 4 | 6319 | 2990 | 1557 | 1364 | ACACAAAGAC…. | ….GUCUUUGUGU |
* RNA2 in the case of two-segmented viruses, i.e. coguviruses, an entovirus, ixoviruses, and a lentinuvirus
ND – not determined
Proteins
Most phenuivirids encode four structural proteins, a large protein (L) encoded by the L segment or RNA1, two external glycoproteins (Gn and Gc) (not encoded by all members) encoded by the M segment, and a nucleocapsid protein (N) encoded by the S segment or RNA3 (RNA2). Additionally, non-structural proteins are expressed from the M segment or RNA2 of members of the genera Coguvirus, Bocivirus, Entovirus, Laulavirus, Lentinuvirus, Mechlorovirus, Phlebovirus, Rubodvirus and Tenuivirus, from the S segment or RNA3 of members of the genera Bandavirus, Mechlorovirus, Mobuvirus, Phlebovirus, Tenuivirus and Uukuvirus, from the S2 segment or RNA4 of members of the genera Horwuvirus, Tenuivirus and Wenrivirus, from additional fifth or sixth RNAs of members of the genera Tenuivirus and Mechlorovirus, and from the seventh and eighth RNAs of the member of the genus Mechlorovirus (Table 3 Phenuiviridae and Figure 2 Phenuiviridae).
Table 3 Phenuiviridae. Proteins encoded by phenuivirids and their approximate masses (kDa)a
Genus | RNA segments | ||||||
| L or RNA1 | M or RNA2 | S or RNA3 (RNA2)a | S2 or RNA4 | |||
| RdRP | NSm or NSvc2 | Gn | Gc | N | NSs or NSv3 | NSs2 or NSvc4 |
Bandavirus | 235–237 | none | 47–64 | 54–56 | 27–28 | 31–36 | none |
Beidivirus | 250 | none | 79 | 56 | 33 | none | none |
| Bocivirus | 252–254 | 53 | 38–39 | none | none | ||
Citricivirus | 253 | none | 56 | 57 | 34 | none | none |
Coguvirus | 250–252 | none | 39–42 | 45–53 | none | ||
Entovirus | 271 | none | 39 | 45 | none | ||
Goukovirus | 238–240 | none | 53–56 | 52–54 | 28–29 | none | none |
Horwuvirus | 332–363 | none | 17–33 | 52–77 | 35–52 | none | 33–41 |
Hudivirus | 255 | none | 84 | 56 | 31 | none | none |
Hudovirus | 275 | none | 107 | 55 | 31 | none | none |
Ixovirus | 250–253 | none | 44–58 | none | none | ||
Laulavirus | 256–275 | 77–79 | none | 28–30 | none | none | |
Lentinuvirus | 367 | none | 35 | 48 | none | ||
Mechlorovirusc | 338–341 | 15–21 | 38–43 | 27–33 | 15–23 | 33–44 | |
Mobuvirus | 256–273 | none | 64–68 | 35–48 | none or 17 | none | |
Phasivirus | 251–262 | none | 72–90 | 53–56 | 29–50 | none | none |
Phlebovirus | 228–242 | none or 14–56 | 47–79 | 45–65 | 16–37 | 16–39 | none |
Pidchovirus | 263–267 | none | 65–73 | 55–56 | 30 | none | none |
Rubodvirus | 276–304 | 44–45 | none | 32 | none | none | |
Tanzavirus | 248 | none | 76 | 54 | 28 | none | none |
Tenuivirusd | 336–341 | 32–36 | 91–109 | 33–36 | 22–24 | 32–33 | |
Uukuvirus | 240–250 | none | none or 54–59 | none or 55 | 27–51 | none or 11–35 | none |
Wenrivirus | 233 | none | 48 | 52 | 27 | none | 46 |
a The proteins of the genera Bandavirus, Goukovirus, Phasivirus, Phlebovirus and Tenuivirus have been demonstrated experimentally to be expressed, while others are only predicted.
b The nucleocapsid proteins are encoded on RNA2 in the case of two segmented viruses, i. e., members of the genera Coguvirus, Entovirus, Ixovirus and Lentinuvirus, and RNA4 or RNA5 in the case of members of the genus Mechlorovirus.
c Members of the genus Mechlorovirus additionally have two or four RNA segments and two or seven proteins with unknown function.
d Some members of the genus Tenuivirus additionally have one or two RNA segments and one or six proteins with unknown function.
Lipids
Enveloped virions contain 20–30% lipids by weight. Lipids are derived from the cellular membranes from which viruses mature and include phopholipids, sterols, fatty acids, and glycolipids. Non-enveloped virions do not contain lipids.
Carbohydrates
Enveloped virions contain 2–7% carbohydrate by weight. Asparagine-linked sugars on the Gn and Gc proteins are largely of the high mannose type when viruses are grown in vertebrate cells. Non-enveloped virions do not contain carbohydrates.
Genome organization and replication
The general genome organization for viruses of each genus is shown in Figure 2 Phenuiviridae. The viral genome is usually comprised of three molecules designated the large segment (L segment or RNA1), the medium segment (M segment or RNA2), and the small segment (S segment or RNA3). Some phenuivirids have up to five additional segments. For almost all phenuivirids, the L segment or RNA1 is a negative-sense RNA and encodes the L protein on the virus-complimentary (vc)RNA. The M segment or RNA2 is a negative-sense RNA and encodes the external Gn and Gc proteins or a non-structural MP (bocaviruses, laulaviruses and rubodviruses). Phleboviruses also encode a non-structural protein (NSm) in a pre-glycoprotein coding region which can act as an apoptosis antagonist. In the case of mechloroviruses and tenuiviruses, RNA2 exhibits an ambisense coding strategy. Tenuivirus RNA2 encodes a glycoprotein-like non-structural protein (NSvc2) on the vcRNA and a viral suppressor of RNA silencing (VSR) that counteracts plant antiviral defence mechanisms based on RNA silencing of the vRNA. Mechlorovirus RNA2 also encodes two non-structural protein of unknown function. S segment or RNA3, or RNA2 in the case of two-segmented viruses, i.e., coguviruses, entoviruses, ixoviruses and lentinuviruses, encodes N. In addition, the S segment or RNA3 of coguviruses, bandviruses, mobuviruses, phleboviruses, tenuiviruses and uukuviruses encodes a non-structural protein on the vRNA. The phlebovirus non-structural protein acts as an interferon antagonist. The tenuivirus non-structural protein (NSv3) shows activity of VSR. Horwuviruses, tenuiviruses, and wenriviruses have a fourth RNA, designated as the S2 segment or RNA4. The horwuvirus and wenrivirus S2 segment is a negative-sense RNA and encodes a non-structural protein of unknown function. The tenuivirus RNA4 is an ambisense RNA and encodes two non-structural proteins, the MP of plant viruses and a major non-capsid protein (NCP) that accumulates in large amounts in the cells of infected plants and vector insects and is presumably associated with symptom development of infected plants. The mechlorovirus segments RNA5 to RNA8 are negative-sense or ambisense and encode non-structural proteins of unknown function.
All replication processes of phenuivirids, i.e., cellular attachment, entry into host cells, transportation to endosomes, penetrating the cytosol, transcription, translation, replication, virion assembly and virion release, occur in and from the cytoplasm. Cellular attachment of enveloped phenuivirid particles is mediated by an interaction of viral glycoproteins with host cell receptors and attachment factors. Dendritic cell‐specific intercellular adhesion molecule‐3‐grabbing nonintegrin (DC‐SIGN) and liver‐specific intercellular adhesion molecule‐3‐grabbing nonintegrin (L‐SIGN) have been reported as the host cell receptor and attachment factor, respectively. Following attachment, the virion enters the cell via clathrin-mediated or caveolin‐mediated endocytosis. Phenuivirids are trafficked from the periphery into the centre of the cytosol via endosomes along intracellular microtubules. During trafficking, early endosomes exchange various materials with the outside and become more acidic. The acidification leads to pH-induced insertion of the viral Gc fusion peptide into the endosomal membranes. The fusion of viral envelopes and endosomal membranes releases the viral RNP complexes into the cytoplasm for replication and transcription of the viral genome. Transcription of genome vRNA into mRNA starts with a "cap‐snatching mechanism", in which host mRNAs are cleaved at a position close to the 5′ cap, mediated by the endonuclease domain of the L protein, as is typical for bunyavirals. Different from many other negative‐sense RNA viruses that terminate transcription at short poly (U) stretches, phenuivirid genomes do not contain poly(U) sequences and transcription stops at termination signal sequences in the untranslated regions of the genome RNAs. Consequently, mRNAs are not polyadenylated. Replication of the virus-sense genome RNA into virus-complementary RNA starts at the 3′-end of the viral genomic RNA and the RNA disengages from the N protein while replication progresses. Both ends of the synthesized RNA bind to specific sites of L and are encapsulated with N and form the progeny complementary RNPs that are used as the template to synthesize the progeny viral genomic RNAs. The progeny viral genomic RNP complexes in the cytoplasm are transported into the lipid bilayer membrane of the Golgi apparatus. The translated precursor glycoproteins are cleaved into Gn and Gc. After Gn and Gc are modified by N-linked glycans and form heterodimers at the endoplasmic reticulum, Gn and Gc are targeted to, and retained in the Golgi apparatus, where virions assemble and bud. Virions bud into Golgi cisternae and are transported to the cell surface by the secretory pathway.
Non-enveloped tenuiviruses do not have glycoprotein genes. Their nonstructural NSvc2 proteins encoded on the second vcRNA2 play roles in cellular attachment, entry into host cells, transportation to endosomes, penetrating the cytosol similar to the glycoproteins of enveloped phenuivirids. That is, the protein is cleaved into two proteins, NSvc2-N and NSvc2-C that interact with the viral RNP complexes. The NSvc2-N binds to an unknown receptor of the vector cells, and binding leads to endocytosis followed by entry of the viral RNPs/NSvc2 complexes into endosomes. Under acidic conditions inside the late endosomes of vector cells, NSvc2‐C undergoes a conformational change that triggers cell membrane fusion, which releases viral RNPs/NSvc2‐N complexes into the cytosol. Transcription of genomic vRNA into mRNA starts with a "cap‐snatching mechanism". The mRNAs of tenuiviruses are not polyadenylated and are truncated relative to the vRNA, similar to the phenuivirids with envelopes. In the case of other non-enveloped phenuivirids, viral entry into host cells is presumed to occur through endocytosis of plant or fungal cells via mechanical wounds, but little is known about the mechanisms of virion attachment, host cell entrance, genome assembly and virion release (Hornak et al., 2016, Ferron et al., 2017, Lu et al., 2019, Koch et al., 2021, Kormelink et al., 2021, Xu et al., 2021, Sun et al., 2022).
 |
| Figure 2 Phenuivridae. Genome organization of members of each genus in the family Phenuiviridae. Coloured boxes depict ORFs that encode N, nucleocapsid protein; Gn and Gc, external glycoproteins; and L, large protein. White boxes depict ORFs that encode MP, non-structural cell-to-cell movement protein; NCP, major non-capsid protein; NSm or NSvc2, non-structural protein on the M segment or RNA2; NSs, non-structural protein on the S segment; and VSR, viral suppressor of RNA silencing. |
Biology
Phenuivirids are ecologically diverse, infecting animals including livestock, humans, birds, crustaceans, plants or fungi. Phenuivirids are also found in invertebrates, including arthropods, some of which may serve as unique hosts or may act as biological vectors for transmission to other animals or plants. Phenuivirids include important pathogens of humans, livestock, seafoods and agricultural crops. Phenuivirids are capable of alternately replicating in their vertebrate or plant hosts, and in their arthropod vectors; they are generally are cytolytic for their hosts but cause little or no cytopathogenicity in their arthropod hosts. Different viruses are transmitted by phlebotomine sand-flies, ticks, mosquitoes, plant hoppers or other arthropod vectors. Some viruses have a very narrow host range, especially for arthropod vectors. Transovarial and venereal transmission have been demonstrated for some phenuivirids in their arthropod vectors. In some instances, avian host and/or vector movements may result in virus dissemination. Some viruses cause a reduction in host-cell protein synthesis in vertebrate cells. In natural infections of mammals, viruses are often targeted to a particular organ or cell type. Some viruses induce cell fusion at low pH. Some members have ion-dependent hemagglutinating activity. Genetic reassortment has been demonstrated for certain members both in vitro and in vivo.
Antigenicity
One or both of the envelope glycoproteins have hemagglutinating and neutralizing antigenic properties. Complement-fixing antigenic determinants are principally associated with nucleocapsid protein.
Derivation of names
Bandavirus: from Bhanjanagar, India, where Bhanja virus was first discovered and, Dàbié Mountains, China, where severe fever with thrombocytopenia syndrome virus was first discovered.
Beidivirus: from Húběi (湖北省), China, where Húběi diptera virus 3 was discovered and Diptera, the order of the fly host.
Bocivirus: from the host fungus species Botrytis cinerea Pers. (1794) in which Botrytis cinerea bocivirus 1 was first discovered.
Citricivirus: from the host aphid species Toxoptera citricida (Kirkaldy, 1907) in which Aphis citricidus bunyavirus was first discovered.
Coguvirus: from citrus concave gum disease, presumed to be caused by citrus concave gum-associated virus.
Entovirus: from Entoleuca, the genus of the host fungus.
Goukovirus: from Gouléako, Côte d'Ivoire, where goukovirus-positive mosquitoes were obtained.
Horwuvirus: from the host horsefly and Wǔhàn (武汉市), China, where Wǔhàn horsefly virus was discovered.
Hudivirus: from Húběi (湖北省), China, where Húběi diptera virus 4 was discovered and Diptera, the order of the fly host.
Hudovirus: from Húběi (湖北省), China, where Húběi lepidoptera virus 1 was discovered and Lepidoptera, the order of the butterfly host.
Ixovirus: from the host tick species Ixodes scapularis (Say, 1821) and I. ricinus (Linnaeus, 1758).
Laulavirus: from Laurel Lake, USA, where Laurel Lake virus was discovered.
Lentinuvirus: from Lentinula edodes ((Berk.) Pegler, 1976), the host of Lentinula edodes negative-strand RNA virus 2.
Mechlorovirus: from melon chlorotic spot disease, caused by melon chlorotic spot virus.
Mobuvirus: from Mothra virus and Bunyavirales.
Phasivirus: from Phasi Charoen, Thailand, where phasivirus-positive mosquitoes were obtained.
Phlebovirus: from phlebotomine vectors of sandfly fever group viruses; in turn derived from the Greek φλέψ (phléps) meaning “vein”.
Pidchovirus: from Pidgey virus and Choristoneura rosaceana (Harris, 1841), the host moth.
Rubodvirus: from apple rubbery wood disease, caused presumably by apple rubbery wood viruses 1 and 2.
Tanzavirus: from Tanzania, where Dar es Salaam virus was discovered.
Tenuivirus: from the Latin tenuis, “thin, fine, weak”, describing the virion morphology of viruses in the genus.
Uukuvirus: from Uukuniemi, a former municipality of Finland, where Uukuniemi virus, assigned to the species Uukuniemi uukuvirus was first isolated.
Wenrivirus: from Wēnzhōu shrimp virus 1, a junior synonym of Mourilyan virus.
Genus demarcation criteria
The coding-complete sequences of all genome segments may be sufficient for phenuivirus classification. Demarcation of genera is based upon comprehensive considerations of virus phylogenetic relationships, genome organization and ecology (e.g., host range, pathobiology and transmission patterns, etc.).
Species demarcation criteria
The criteria demarcating species in the genus are:
• Viruses assigned to different species have less than 95% identity in the amino acid sequence of the RdRP
Relationships within the family
Viruses within the same genus form monophyletic clades based upon phylogenetic reconstruction using the L protein sequence (Figure 3 Phenuiviridae).
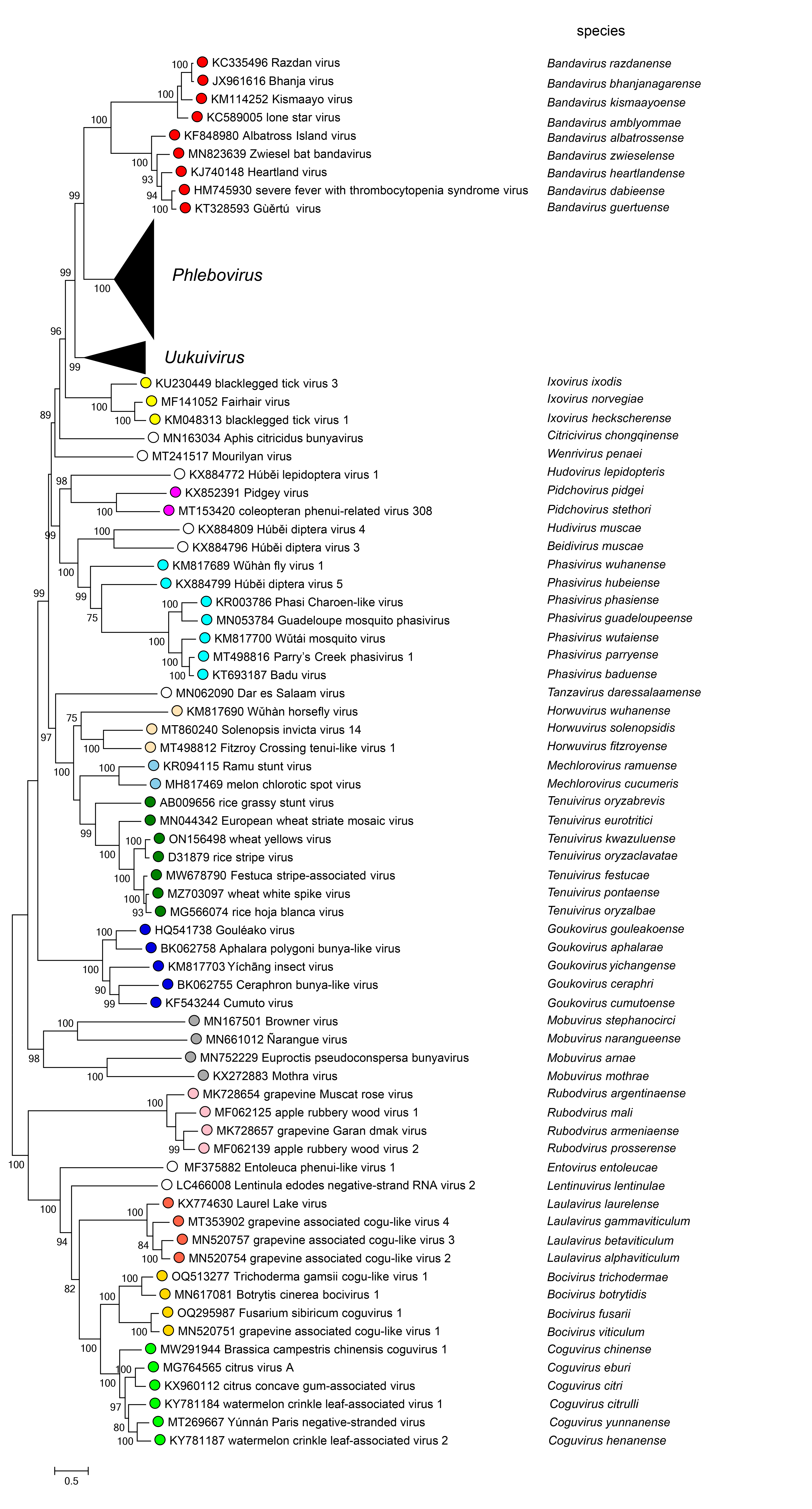 |
| Figure 3 Phenuiviridae Phylogenetic analysis of phenuivirid genera. Phylogenetic reconstruction is based on a MAFFT-alignment of the RdRP amino acid sequences of phenuivirids using E-INS algorithm. The ML phylogenetic tree was inferred using RaxML-NG performing 1,000 bootstrap replicates. Trees were inferred under the WAG substitution model. Tree branches are proportional to genetic distances between sequences and the scale bars at the bottom indicate substitutions per amino acid. The branches including phleboviruses and uukuviruses are collapsed for clarity. Figures with these regions expanded are shown on the Phlebovirus genus page as Figure 1 Phlebovirus and on the Uukuvirus genus page as Figure 1 Uukuvirus. |
Relationships with other taxa
Phenuivirids are closely related to viruses in the bunyaviral families Arenaviridae, Discoviridae, Leishbuviridae, Mypoviridae, Nairoviridae and Wupedeviridae, and more distantly related to viruses in the bunyaviral families Cruliviridae, Fimoviridae, Peribunyaviridae, Phasmaviridae, Tospoviridae and Tulasviridae.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Alternaria tenuissima negative-stranded RNA virus 2 | L: MK584855; M: BK061363; S: BK061364 | AtNSRV2 |
| Austropotamobius brown spot virus (=bunya-like brown spot virus) | L: MK881590; M: MK881591; S: MK881592 | AuBSV |
| Beidivirus atrichopogon | L: BK063256; M: BK6063257; S: BK063258 | BVa |
| Culex bunyavirus 1 | L: MH188051 | CuBV1 |
| Fusarium asiaticum mycobunyavirus 1 | L: MZ969068; M: MZ969069; S: MZ969070 | FaMBV1 |
| Fusarium poae negative-stranded RNA virus 2 | L: LC150619 | FpNSV2 |
| Húběi bunya-like virus 2 | Seg1: KX884780; Seg2: KX884781 | HbBLV2 |
| Húběi bunya-like virus 3 | Seg1: KX884778; Seg2: KX884779 | HbBLV3 |
| Húběi bunya-like virus 13 | Seg1: KX884784; Seg2: KX884785 | HbBLV13 |
| Húběi insect virus 1 | Seg1: KX884735; Seg2: KX884736 | HbIV1 |
| lettuce dieback associated virus | RNA1: MT386514; RNA2: MT386515; RNA3: MT386516 | LeDaV |
| Linepithema humile bunyan-like virus 1 | L: MH213237; S: MH213238 | LBLV1 |
| otter fecal phlebovirus | M: KF823817*; S: KF823816* | OFPV |
| Pälkäne phenui-like virus 2 | L: ON955229 | PaPLV2 |
| salarivirus Mos8CM0 | L: KX924627*; M: KX924628*; S: KX924629 | SaV |
| Sclerotinia sclerotiorum negative-stranded RNA virus 5 | L: KF913892; M: BK061361; S: BK061362 | SsNSRV5 |
| Sclerotinia sclerotiorum phlebo-like virus 1 | L: KC601996 | SsNSRV1 |
| Shāhé heteroptera virus 3 | L: KX884813; M: KX884814; S: KX884815 | ShaHV3 |
| Shāyáng bunya-like virus 1 | Seg1: KX884835 | ShBLV |
| Shuāngào insect virus 3 | L: KM817681; M: KM817716; S: KM817742 | ShIV3 |
| soybean cyst nematode associated rice stripe virus | L: HM849041 | SCNaRSV |
| soybean cyst nematode associated Uukuniemi virus | L: HM849040 | SCNaUV |
| soybean thrips-associated tenui-like virus 1 | RNA1: MT224144; RNA?: MW033650 | STaTlV1 |
| sugar beet cyst nematode virus 2 | L: KY609505; M: MH282849* | SBCNV2 |
| Timbillica virus | L: MK026589; S: MK026590* | TimV |
| Vietnamese bat bunyavirus | L: KX886759; S: KX886760 | ViBBV |
| Wēnlǐng crustacean virus 7 | Seg1: KX884859; Seg2: KX884860 | WeCV7 |
| Wǔhàn insect virus | L: KM817691; S: KM817752* | WuIV |
| Wǔhàn insect virus 16 | Seg1: KX884854; Seg2: KX884855 | WuIV16 |
| Xīnzhōu bunya-like virus 1 | Seg1: KX884868 | XBLV1 |
| Zhee mosquito virus | L: KM817705 | ZhMV |
* Sequences do not comprise the complete genome segment.
Virus names and virus abbreviations are not official ICTV designations.

