Family: Geminiviridae
Genus: Mastrevirus
Distinguishing features
Mastreviruses have a monopartite genome, are leafhopper-transmitted that have only been identified in plants occurring in the Old World (Shepherd et al., 2010). They infect tropical and temperate cereals and some vegetable crops. Many cereal-infecting mastreviruses also infect wild grasses, which are often considered to be the natural hosts of these viruses. This group of geminiviruses utilizes transcript splicing, and therefore their genomes contain introns. The proteins translated from the spliced mRNAs encoded by C1 and C2 ORFs are involved in replication and encode the replication initiator protein. In addition, regulation of virion-sense gene expression in some grass-infecting mastreviruses occurs by differential splicing of two virion sense transcripts transcribed from the V1 and V2 ORFs.
Virion
See discussion under family description.
Genome organization and replication
The mastrevirus genome consists of a single component of circular ssDNA of 2.6–2.8 kb. A small complementary-sense DNA containing 5′-ribonucleotides, annealed within the small intergenic region to the genomic DNA of Chloris striate mosaic virus (CSMV), Digitaria streak virus (DSV), maize streak virus (MSV), tobacco yellow dwarf virus (TYDV) and wheat dwarf virus (WDV), may be involved in priming complementary-sense DNA synthesis. The small, annealed DNA is subsequently encapsidated with genomic ssDNA.
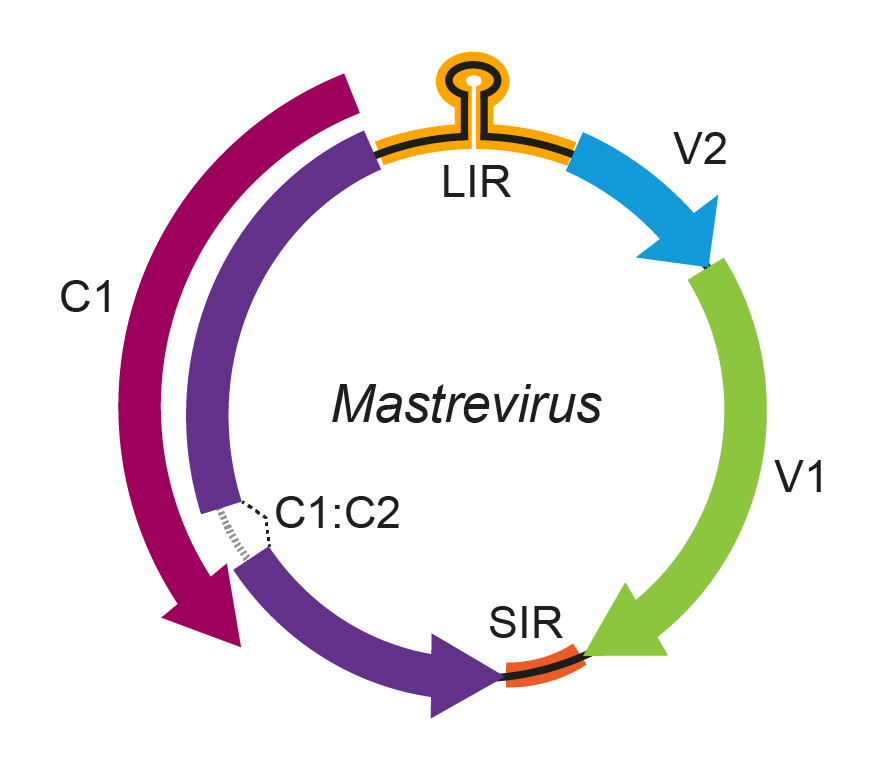
Mastrevirus genomes encode four proteins (Figure 1. Mastrevirus). The two encoded on the virion-sense strand are the coat protein (CP, gene V1) that encapsidates the virion-sense ssDNA and acts as a nuclear shuttle protein (NSP) for viral DNA, and the movement protein (MP, gene V2), that functions in cell-to-cell movement. The CP appears to regulate the balance of ssDNA and dsDNA accumulation. Regulation of virion-sense gene expression in MSV and grass-infecting relatives occurs by differential transcript splicing. The complementary-sense strand encodes the replication-associated protein (Rep), expressed from genes C1 and C2 by transcript splicing. The Rep protein initiates rolling circle replication by introducing a nick into the conserved 5′-TAATATTAC-3′ sequence in the virion-sense strand. Rep binds to the large subunit of the replication factor C clamp loader complex, suggesting a role in the recruitment of host replication factors to the origin of replication. The RepA protein (ORF C1), also encoded on the complementary-sense strand, binds to the plant homologue of retinoblastoma protein (Rb) to regulate cell-cycle progression, altering the environment of terminally differentiated cells to provide host factors that support viral DNA replication. Both Rep and RepA bind to the origin of replication as multimeric proteins.
|
|
|
Figure 1. Mastrevirus. Genomic organization of mastreviruses. ORFs are denoted as being encoded on the virion-sense (V) or complementary-sense (C) strand, and corresponding protein products are indicated. The position of the stem-loop motif containing the conserved 5′-TAATATTAC-3′ sequence in the long intergenic region (LIR) is shown. Introns (open boxes) occur in ORF V2 of some mastreviruses and between ORFs C1 and C2 of all mastreviruses. CP, coat protein; MP, movement protein; Rep, replication-associated protein. |
The genome contains two intergenic regions. The long intergenic region (LIR) is the location of both the v-ori and transcription start sites, while the short intergenic region (SIR) is the location of both the complementary-strand replication origin and transcription termination sites.
Biology
Host range
The host range of mastreviruses is relatively narrow. With the exception of TYDV and chickpea chlorotic dwarf virus (CpCDV), which infect plants in the families Solanaceae and Fabaceae, respectively, the host range of mastreviruses is limited to monocot plant species.
Transmission
Mastreviruses are transmitted by specific leafhoppers (order Hemiptera, family Cicadellidae), in most cases by a single species. In common with all geminiviruses, transmission is persistent, circulative and non-propagative. Mastreviruses are normally not transmissible by mechanical inoculation, although MSV has been transmitted experimentally via a vascular puncture technique using maize seeds. Most members are transmitted experimentally to plants by Agrobacterium-mediated transfer (agroinoculation) from partially or tandemly repeated cloned genomic DNA.
Antigenicity
Serological analyses indicate that grass-infecting mastreviruses from the same continent constitute distinct groupings: there is an African streak virus group [MSV, Panicum streak virus (PanSV), sugarcane streak virus (SSV), sugarcane streak Egypt virus (SSEV) and sugarcane streak Reunion virus (SSREV)], an Australasian striate mosaic virus group [Bromus catharticus striate mosaic virus (BCSMV), CSMV and Paspalum striate mosaic virus (PSMV)], and the very distinct Asian Miscanthus streak virus (MiSV) and European WDV. Although DSV originates from Vanuatu, it is most closely related to the African mastreviruses. Grass geminiviruses originating from different continents are antigenically either unrelated or only distantly related. Mastreviruses that infect dicot plants are not antigenically related to those that infect monocotyledonous plants.
Species demarcation criteria
The following criteria are used as a guideline to establish taxonomic status:
- Nucleotide sequence identity: isolates with a full-length nucleotide sequence identity of <78% compared to the isolates of any recognized species are considered to belong to a distinct species. This primary criterion can be applied in the absence of additional data, as long as sequence identities are calculated using true pairwise alignments with pairwise deletion of gaps. Nonetheless, when approaching the cut-off value, the biological properties of the virus should also be taken into account.
- Trans-replication of genomic components: the inability of a Rep protein of one virus to trans-replicate the genomic component of another virus suggests the two viruses should be considered members of different species.
- Coat protein: members of different species may display serological differences.
- Different vector species: viruses that are not transmissible by the same insect species may be considered to be belong to different species.
- Natural host range and symptom phenotype: these characteristics may differ between members of different species, but their commonest use will be to distinguish between the strains of individual species.
A strain demarcation threshold has been established by the Geminiviridae Study Group at <94% shared nt identity of full length nucleotide sequences.